7QYJ
 
 | | The structure of T. forsythia NanH | | Descriptor: | BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7MY9
 
 | | Structure of proline utilization A with 1,3-dithiolane-2-carboxylate bound in the proline dehydrogenase active site | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Campbell, A.C. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.
Acs Chem.Biol., 16, 2021
|
|
7R56
 
 | |
6GYH
 
 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
7QY9
 
 | | The structure of T.forsythia NanH with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
8AHC
 
 | | Crystal structure of the BRD9 bromodomain with BI-7189 | | Descriptor: | Bromodomain-containing protein 9, [2,6-dimethoxy-4-(1,2,5-trimethyl-6-oxidanylidene-pyridin-3-yl)phenyl]methyl-dimethyl-azanium | | Authors: | Bader, G, Boettcher, J, Weiss-Puxbaum, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
7QYP
 
 | | The structure of T. forsythia NanH | | Descriptor: | 1,2-ETHANEDIOL, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7NMK
 
 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis with bound methylation product 1-methoxyquinolin-4(1H)-one | | Descriptor: | 1-methoxy-4-oxoquinoline, 2-heptyl-1-hydroxyquinolin-4(1H)-one methyltransferase, FORMIC ACID, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
8AIN
 
 | | MCUGI SAUNG complex | | Descriptor: | 1,2-ETHANEDIOL, MCUGI, SULFATE ION, ... | | Authors: | Muselmani, W, Savva, R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
7QZ3
 
 | | The structure of T. forsythia NanH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
8A9O
 
 | | Structure of the polyamine acetyltransferase DpA | | Descriptor: | ACETYL COENZYME *A, BROMIDE ION, COENZYME A, ... | | Authors: | Garcia-Pino, A, Jurenas, D. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | A polyamine acetyltransferase regulates the motility and biofilm formation of Acinetobacter baumannii.
Nat Commun, 14, 2023
|
|
7MYA
 
 | |
8AIM
 
 | | Ugi-2 SAUNG complex | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Muselmani, W, Savva, R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
7MYC
 
 | |
6GZW
 
 | |
8AIL
 
 | | Bacillus phage VMY22 p56 in complex with Bacillus weidmannii Ung | | Descriptor: | Bacillus phage VMY22 p56, GLYCEROL, IODIDE ION, ... | | Authors: | Muselmani, W, Bagneris, C, Savva, R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
8A9N
 
 | | Structure of DpA polyamine acetyltransferase in complex with 1,3-DAP | | Descriptor: | 1,3-DIAMINOPROPANE, Acetyltransferase, COENZYME A, ... | | Authors: | Garcia-Pino, A, Jurenas, D. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | A polyamine acetyltransferase regulates the motility and biofilm formation of Acinetobacter baumannii.
Nat Commun, 14, 2023
|
|
6GZS
 
 | |
8AA7
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment | | Descriptor: | 2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Cytochrome P450 protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment
To Be Published
|
|
6GZY
 
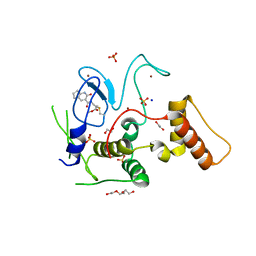 | | HOIP-fragment5 complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF31, SODIUM ION, ... | | Authors: | Johansson, H, Tsai, Y.C.I, Fantom, K, Chung, C.W, Martino, L, House, D, Rittinger, K. | | Deposit date: | 2018-07-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Covalent Ligand Screening Enables Rapid Discovery of Inhibitors for the RBR E3 Ubiquitin Ligase HOIP.
J. Am. Chem. Soc., 141, 2019
|
|
6CPL
 
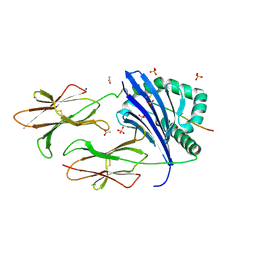 | | Crystal structure of DR11 presenting the gag293 epitope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
8ABO
 
 | |
6H01
 
 | |
8TXN
 
 | |
8ABS
 
 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with testosterone and putative ligand 4,6-dimethyloctanoic acid | | Descriptor: | (4~{S},6~{S})-4,6-dimethyloctanoic acid, Cytochrome P450, HEME B/C, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
