9UL7
 
 | |
9UAV
 
 | |
3OAB
 
 | | Mint deletion mutant of heterotetrameric geranyl pyrophosphate synthase in complex with ligands | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
J.Mol.Biol., 404, 2010
|
|
9OGK
 
 | | Refinement of PDB-3J5R against EMD-8117 using EMAN2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 1 | | Authors: | Chen, M. | | Deposit date: | 2025-04-30 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Building molecular model series from heterogeneous CryoEM structures using Gaussian mixture models and deep neural networks.
Commun Biol, 8, 2025
|
|
9LNX
 
 | |
9LNL
 
 | |
4YXU
 
 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (4). | | Descriptor: | 4-propylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
6QTM
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ty5 pS1095 peptide | | Descriptor: | Regulatory protein SIR4, Ribonuclease H, SULFATE ION | | Authors: | Gut, H, Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
5D4C
 
 | |
9QMP
 
 | | E.coli seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase | | Authors: | Cusack, S. | | Deposit date: | 2025-03-24 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A.
Nature, 347, 1990
|
|
9QOF
 
 | | E.coli seryl-tRNA synthetase (Arm deletion mutant) bound to sulphamoyl seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cusack, S, Belrhali, H, Price, S, Leberman, R. | | Deposit date: | 2025-03-26 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Seryl-tRNA synthetase from Escherichia coli,implication of its N-terminal domain in aminoacylation activity and specificity.
Nucleic Acids Res, 22, 1994
|
|
8J3S
 
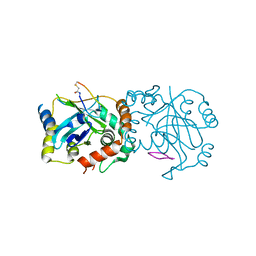 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
4YXO
 
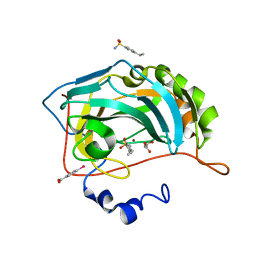 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (3). | | Descriptor: | 4-ethylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
3WK2
 
 | |
2P8P
 
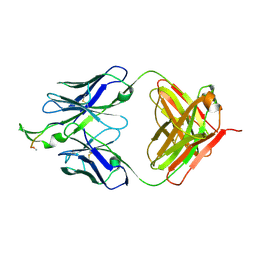 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LELDKWASLW[N-Ac] | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
4Q2Y
 
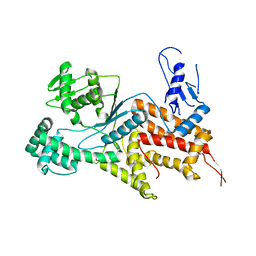 | | Crystal structure of Arginyl-tRNA synthetase | | Descriptor: | Arginine--tRNA ligase, cytoplasmic | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
1TY9
 
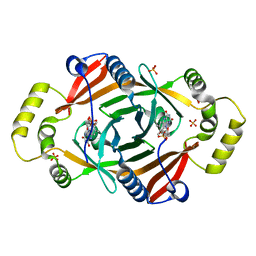 | | X-RAY CRYSTAL STRUCTURE OF PHZG FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1LY3
 
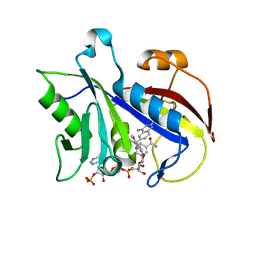 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2Z4J
 
 | |
4QH4
 
 | |
2P8L
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Julien, J.P, Bryson, S, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
5NY6
 
 | | Carbonic Anhydrase II Inhibitor RA12 | | Descriptor: | 4-chloranyl-~{N}-[(1~{R})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, 4-chloranyl-~{N}-[(1~{S})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
2ONE
 
 | |
5UJJ
 
 | |
4Q2T
 
 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-arginine | | Descriptor: | ARGININE, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
