9G31
 
 | |
4Q4O
 
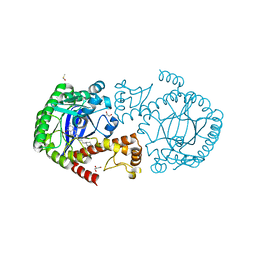 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 6-Amino-2-{[2-(piperidin-1-yl)ethyl]amino}-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-{[2-(piperidin-1-yl)ethyl]amino}-3,5-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Beyond Affinity: Enthalpy-Entropy Factorization Unravels Complexity of a Flat Structure-Activity Relationship for Inhibition of a tRNA-Modifying Enzyme.
J.Med.Chem., 57, 2014
|
|
9GDL
 
 | |
9GDN
 
 | |
9GDU
 
 | |
9GE9
 
 | |
6PAB
 
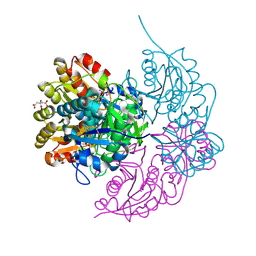 | |
9G2O
 
 | |
9GDT
 
 | |
9GE1
 
 | |
9GEC
 
 | |
5OAL
 
 | | Crystal structure of mutant AChBP in complex with strychnine (T53F, Q74R, Y110A, I135S, G162E) | | Descriptor: | ARGININE, STRYCHNINE, Soluble acetylcholine receptor | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-22 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
6X83
 
 | |
6PAA
 
 | |
6X82
 
 | | Crystal Structure of TNFalpha with isoquinoline compound 4 | | Descriptor: | 8-[4-(2-{5-[(4-methylpiperazin-1-yl)methyl]-2-(1H-pyrrolo[3,2-c]pyridin-3-yl)phenoxy}ethyl)phenyl]isoquinoline, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
6X86
 
 | | Crystal Structure of TNFalpha with indolinone compound 11 | | Descriptor: | 3-[(6-{2-[(3R)-4-(hydroxyacetyl)-3-methylpiperazin-1-yl]pyrimidin-5-yl}-2,2-dimethyl-3-oxo-2,3-dihydro-1H-indol-1-yl)methyl]pyridine-2-carbonitrile, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
5OA0
 
 | | Crystal structure of mutant AChBP in complex with strychnine (T53F, Q74R, Y110A, I135S, W164F) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, STRYCHNINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
6PA6
 
 | |
5H4V
 
 | |
8USW
 
 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
8PPK
 
 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
6M0T
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin derivative (CL-2) | | Descriptor: | (3R)-3-[(R)-[(2R,6S)-6-methyloxan-2-yl]-oxidanyl-methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y. | | Deposit date: | 2020-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Cladosporin-Based Inhibitors of Malaria Parasites.
Acs Infect Dis., 7, 2021
|
|
8SLY
 
 | | Rat TRPV2 bound with 2 CBD ligands in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 2, ... | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cannabidiol sensitizes TRPV2 channels to activation by 2-APB.
Elife, 12, 2023
|
|
6L4Q
 
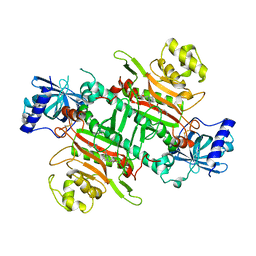 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Clado-B | | Descriptor: | (3R)-3-[[(3R)-3-methylpiperidin-1-yl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y, Mishra, S, Harlos, K. | | Deposit date: | 2019-10-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin inhibitor, Cla-B
Chembiochem, 2021
|
|
6L3Y
 
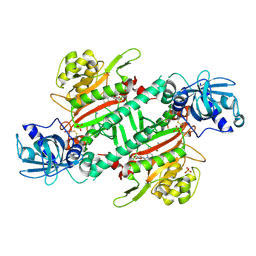 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Clado-C | | Descriptor: | (3R)-3-[[(3S)-3-ethylpiperidin-1-yl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Babbar, P, Sharma, A, Mishra, S, Manickam, Y, Harlos, K. | | Deposit date: | 2019-10-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin inhibitor, Cla-B
Chembiochem, 2021
|
|
