186L
 
 | |
4J0S
 
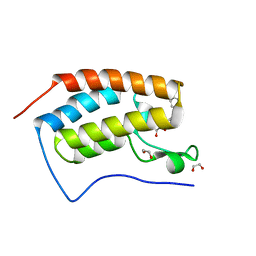 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(S)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
6FSQ
 
 | |
6MYT
 
 | | Avian mitochondrial complex II with Atpenin A5 bound, sidechain in pocket | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYP
 
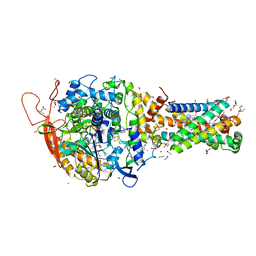 | | Avian mitochondrial complex II with TTFA (thenoyltrifluoroacetone) bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, BICARBONATE ION, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYU
 
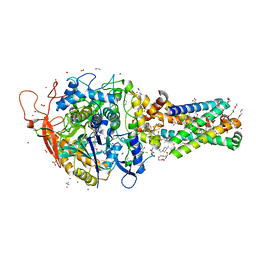 | | Avian mitochondrial complex II crystallized in the presence of HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYO
 
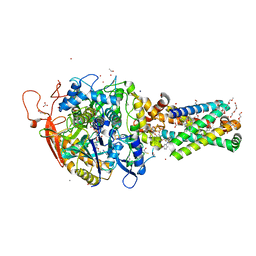 | | Avian mitochondrial complex II with flutolanyl bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6CJT
 
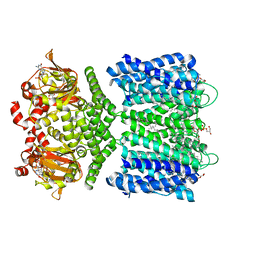 | | Structure of the SthK cyclic nucleotide-gated potassium channel in complex with cGMP | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CYCLIC GUANOSINE MONOPHOSPHATE, SthK cyclic nucleotide-gated potassium channel | | Authors: | Nimigean, C.M, Rheinberger, J. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures.
Elife, 7, 2018
|
|
3SAO
 
 | | The Siderocalin Ex-FABP functions through dual ligand specificities | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2,3-DIHYDROXY-BENZOIC ACID, Extracellular fatty acid-binding protein, ... | | Authors: | Correnti, C, Strong, R.K, Clifton, M.C. | | Deposit date: | 2011-06-03 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Galline Ex-FABP Is an Antibacterial Siderocalin and a Lysophosphatidic Acid Sensor Functioning through Dual Ligand Specificities.
Structure, 19, 2011
|
|
4F27
 
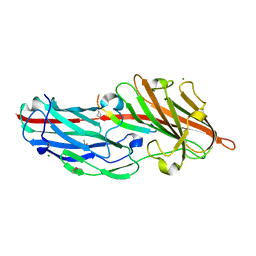 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Fibrinogen alpha chain | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
4F20
 
 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Dermokine | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
5ILC
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound 2, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2016-03-04 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
6HWP
 
 | |
1BTJ
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE, APO FORM, CRYSTAL FORM 2 | | Descriptor: | PROTEIN (SERUM TRANSFERRIN) | | Authors: | Jeffrey, P.D, Bewley, M.C, Macgillivray, R.T.A, Mason, A.B, Woodworth, R.C, Baker, E.N. | | Deposit date: | 1998-09-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin.
Biochemistry, 37, 1998
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
5ILF
 
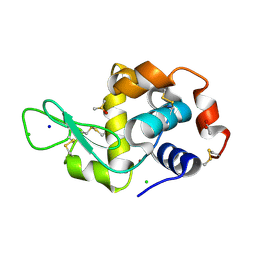 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 4, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2016-03-04 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
1LAH
 
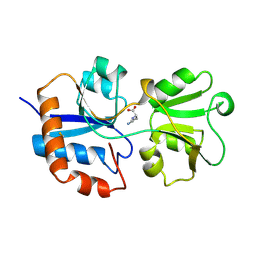 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
6FT5
 
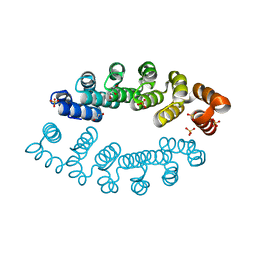 | | Structure of A3_A3, an artificial bi-domain protein based on two identical alphaRep A3 domains | | Descriptor: | GLYCEROL, SULFATE ION, alphaRep A3_A3 | | Authors: | Li de la Sierra-Gallay, I, Leger, C, Di Meo, T. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Ligand-induced conformational switch in an artificial bidomain protein scaffold.
Sci Rep, 9, 2019
|
|
5IHG
 
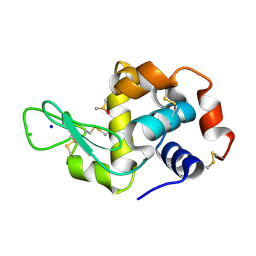 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound I, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-02-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
6W9M
 
 | |
8FGY
 
 | |
8FH0
 
 | |
2JOC
 
 | | Mouse Itch 3rd domain phosphorylated in T30 | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
3S2V
 
 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | Descriptor: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
