7ASL
 
 | |
7NU0
 
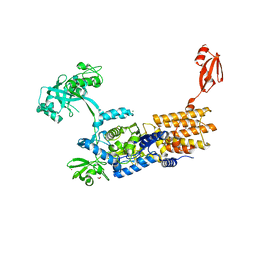 | | Crystal Structure of Neisseria gonorrhoeae LeuRS L550A mutant in Complex with ATP and L-leucinol | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-METHYL-PENTAN-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
8UHS
 
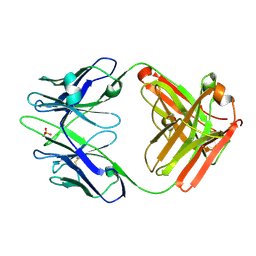 | | anti-Phosphohistidine Fab hSC44.ck.20.elbow bound to phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, hSC44.ck.20.elbow Fab heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
7FZJ
 
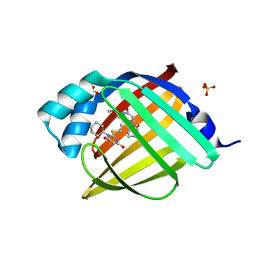 | | Crystal Structure of human FABP4 in complex with 3-cyclohexyl-2-ethylsulfanyl-6-hydroxypyrimidin-4-one | | Descriptor: | 1-cyclohexyl-2-(ethylsulfanyl)pyrimidine-4,6(1H,5H)-dione, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Jin, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6WLZ
 
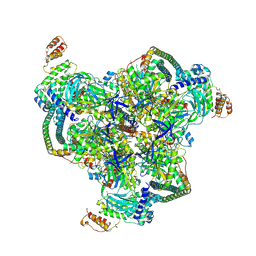 | | The V1 region of human V-ATPase in state 1 (focused refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SidK, V-type proton ATPase catalytic subunit A, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
8T8K
 
 | |
6CPV
 
 | |
4TWS
 
 | | Gadolinium Derivative of Tetragonal Hen Egg-White Lysozyme at 1.45 A Resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Holton, J.M, Classen, S, Frankel, K.A, Tainer, J.A. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures.
Febs J., 281, 2014
|
|
6UA3
 
 | |
6G0S
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SIRT7 peptide (K272ac/K275ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, NAD-dependent protein deacetylase sirtuin-7 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
7UOS
 
 | | Structure of WNK1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 4-[bromo(dichloro)methanesulfonyl]-N-cyclohexyl-2-nitroaniline, Serine/threonine-protein kinase WNK1 | | Authors: | Akella, R, Goldsmith, E.J, Akella, R. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of WNK1 inhibitor complex
To Be Published
|
|
8E4Y
 
 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase 1, mitochondrial, ... | | Authors: | Johnson, Z.L, Wasilko, D.J, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7FX3
 
 | | Crystal Structure of apo human FABP4, tetragonal form | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein, adipocyte | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8SV9
 
 | | Crystal structure of ULK1 kinase domain with inhibitor MR-2088 | | Descriptor: | (4P)-4-[(2P)-2-(1,2,5,6-tetrahydropyridin-3-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-(2,2,2-trifluoroethyl)thiophene-2-carboxamide, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of potent and selective ULK1/2 inhibitors based on 7-azaindole scaffold with favorable in vivo properties.
Eur.J.Med.Chem., 266, 2024
|
|
4UFM
 
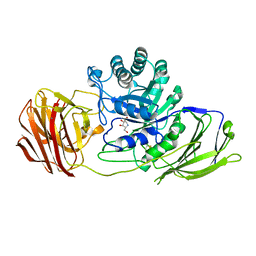 | | Mouse Galactocerebrosidase complexed with 1-deoxy-galacto-nojirimycin DGJ | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hill, C.H, Viuff, A.H, Spratley, S.J, Salamone, S, Christensen, S.H, Read, R.J, Moriarty, N.W, Jensen, H.H, Deane, J.E. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Azasugar Inhibitors as Pharmacological Chaperones for Krabbe Disease.
Chem.Sci., 6, 2015
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
8TD1
 
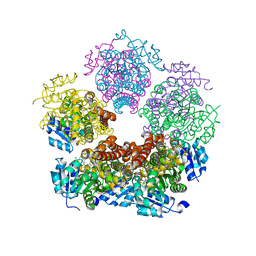 | |
5IJV
 
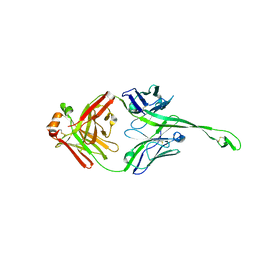 | | Crystal structure of bovine Fab E03 | | Descriptor: | bovine Fab E03 heavy chain, bovine Fab E03 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
7U04
 
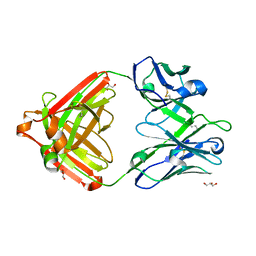 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
8E0E
 
 | | nbF3:CaV beta subunit 2a complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nirwan, N, Minor, D.L. | | Deposit date: | 2022-08-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective posttranslational inhibition of Ca V beta 1 -associated voltage-dependent calcium channels with a functionalized nanobody.
Nat Commun, 13, 2022
|
|
8JJG
 
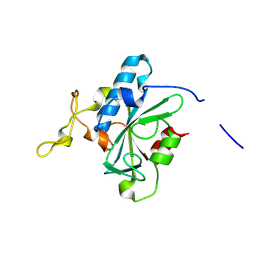 | | Crystal structure of QW-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJI
 
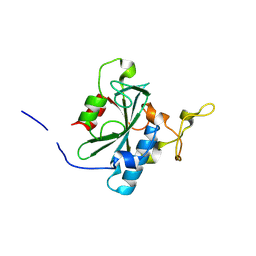 | | Crystal structure of QR-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
6FTC
 
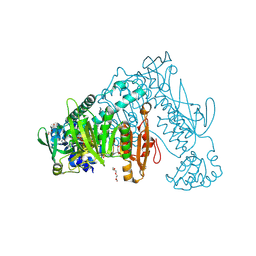 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Silvestri, I, Fata, F, MIele, A.E, Boumis, G, Williams, D.L, Angelucci, F. | | Deposit date: | 2018-02-21 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Discovery of a Regulatory Site in Thioredoxin Glutathione Reductase Acting as "Doorstop" for NADPH Entry.
ACS Chem. Biol., 13, 2018
|
|
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7QIY
 
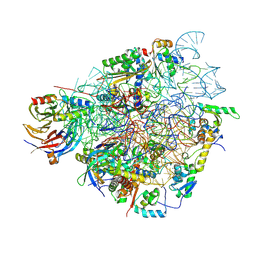 | | Specific features and methylation sites of a plant ribosome. 40S head ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA head, 40S head ribosomal protein eS19, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
