8FMU
 
 | | Crystal structure of human Brachyury G177D variant in complex with SJF-4601 | | Descriptor: | N-(3-chloro-4-fluorophenyl)-3-[4-(dimethylamino)butanamido]-4-methoxybenzamide, T-box transcription factor T | | Authors: | Bebenek, A, Linhares, B, Jaime-Figueroa, S, Butrin, A, Crews, C. | | Deposit date: | 2022-12-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of a Small Molecule Downmodulator for the Transcription Factor Brachyury.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1CNQ
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-6-PHOSPHATE AND ZINC IONS | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE-1,6-BISPHOSPHATASE, PHOSPHATE ION, ... | | Authors: | Choe, J, Poland, B.W, Fromm, H, Honzatko, R. | | Deposit date: | 1999-05-21 | | Release date: | 1999-05-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase.
Biochemistry, 37, 1998
|
|
6XIU
 
 | |
6XIV
 
 | | SeMet-Rns, in complex with potential inhibitor | | Descriptor: | (2S)-2-hydroxybutanedioic acid, DECANOIC ACID, Regulatory protein Rns | | Authors: | Midgett, C.R, Kull, F.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the master regulator Rns reveals an inhibitor of enterotoxigenic Escherichia coli virulence regulons.
Sci Rep, 11, 2021
|
|
1D4R
 
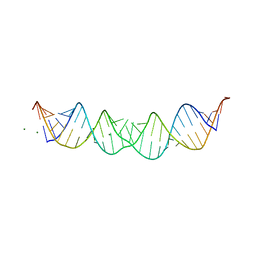 | | 29-mer fragment of human srp rna helix 6 | | Descriptor: | 29-MER OF MODIFIED SRP RNA HELIX 6, MAGNESIUM ION | | Authors: | Wild, K, Weichenrieder, O, Leonard, G.A, Cusack, S. | | Deposit date: | 1999-10-05 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2 A structure of helix 6 of the human signal recognition particle RNA
Structure Fold.Des., 7, 1999
|
|
1CYL
 
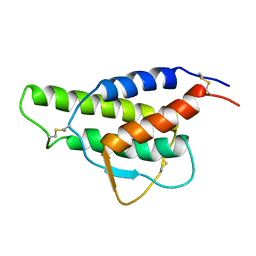 | |
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
1DGM
 
 | | CRYSTAL STRUCTURE OF ADENOSINE KINASE FROM TOXOPLASMA GONDII | | Descriptor: | ACETIC ACID, ADENOSINE, ADENOSINE KINASE, ... | | Authors: | Cook, W.J, DeLucas, L.J, Chattopadhyay, D. | | Deposit date: | 1999-11-24 | | Release date: | 2000-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenosine kinase from Toxoplasma gondii at 1.8 A resolution.
Protein Sci., 9, 2000
|
|
1DOR
 
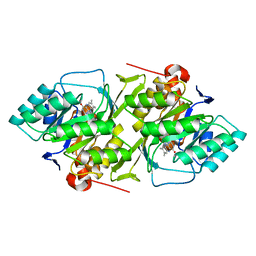 | | DIHYDROOROTATE DEHYDROGENASE A FROM LACTOCOCCUS LACTIS | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE | | Authors: | Rowland, P, Larsen, S. | | Deposit date: | 1997-01-14 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the flavin containing enzyme dihydroorotate dehydrogenase A from Lactococcus lactis.
Structure, 5, 1997
|
|
1DUH
 
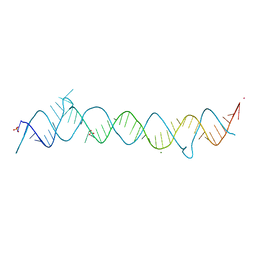 | | CRYSTAL STRUCTURE OF THE CONSERVED DOMAIN IV OF E. COLI 4.5S RNA | | Descriptor: | 4.5S RNA DOMAIN IV, LUTETIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Jovine, L, Hainzl, T, Oubridge, C, Scott, W.G, Li, J, Sixma, T.K, Wonacott, A, Skarzynski, T, Nagai, K. | | Deposit date: | 2000-01-17 | | Release date: | 2000-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA.
Structure Fold.Des., 8, 2000
|
|
1DUQ
 
 | |
4GWO
 
 | |
7KSO
 
 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6GBZ
 
 | | 50S ribosomal subunit assembly intermediate state 5 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
7KSR
 
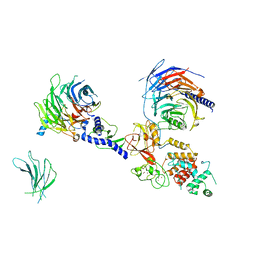 | | PRC2:EZH1_A from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KTP
 
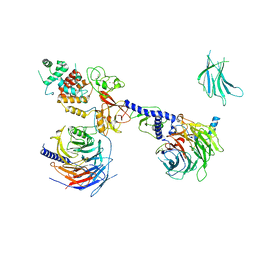 | | PRC2:EZH1_B from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
6GC8
 
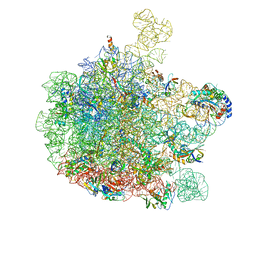 | | 50S ribosomal subunit assembly intermediate - 50S rec* | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GC4
 
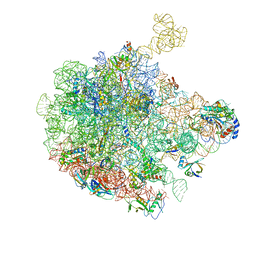 | | 50S ribosomal subunit assembly intermediate state 3 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
8IR1
 
 | | human nuclear pre-60S ribosomal particle - State A | | Descriptor: | 28S rRNA, 5.8S rRMA, 5S RNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
6GC0
 
 | | 50S ribosomal subunit assembly intermediate state 4 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6T9J
 
 | | SAGA Tra1 module | | Descriptor: | Transcription factor SPT20, Transcription initiation factor TFIID subunit 12, Transcription-associated protein 1 | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6GGA
 
 | |
6GGE
 
 | | p53 cancer mutant Y220C in complex with small-molecule stabilizer PK9327 | | Descriptor: | Cellular tumor antigen p53, ZINC ION, [9-ethyl-7-(5-methylthiophen-2-yl)carbazol-3-yl]methyl-methyl-azanium | | Authors: | Joerger, A.C, Bauer, M.R. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25000513 Å) | | Cite: | A structure-guided molecular chaperone approach for restoring the transcriptional activity of the p53 cancer mutant Y220C.
Future Med Chem, 11, 2019
|
|
