5X8F
 
 | |
6PA8
 
 | |
3LGV
 
 | | H198P mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
2R7K
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMPPCP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
8V2W
 
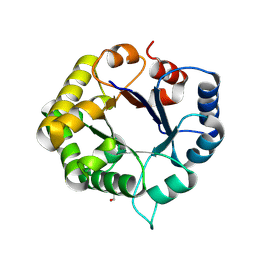 | | Crystal Structure of the ancestral triosephosphate isomerase reconstruction of the last opisthokont common ancestor obtained by Bayesian inference | | Descriptor: | GLYCEROL, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8V0A
 
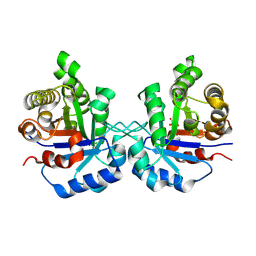 | | Crystal Structure of the worst case of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
2R7M
 
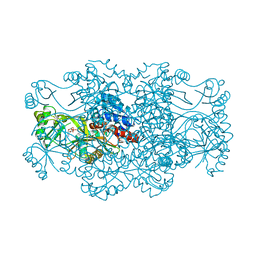 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMP | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
3CGT
 
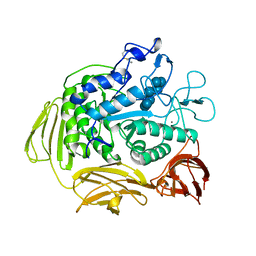 | |
6PAB
 
 | |
3CQT
 
 | | N53I V55L MUTANT of FYN SH3 DOMAIN | | Descriptor: | Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Neculai, A.M, Zarrine-Afsar, A, Howell, P.L, Davidson, A, Chan, H.S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6PA6
 
 | |
4A2H
 
 | | Crystal Structure of Laccase from Coriolopsis gallica pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | De La Mora, E, Valderrama, B, Horjales, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3LGW
 
 | |
8GUH
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Tris | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [4-[[[2-(hydroxymethyl)-1,3-bis(oxidanyl)propan-2-yl]amino]methyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Sphingobacterium multivorum serine palmitoyltransferase complexed with tris(hydroxymethyl)aminomethane.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7P5X
 
 | | Mycobacterial RNAP with transcriptional activator PafBC | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Mueller, A.U, Kummer, E, Schilling, C.M, Ban, N, Weber-Ban, E. | | Deposit date: | 2021-07-15 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transcriptional control of mycobacterial DNA damage response by sigma adaptation.
Sci Adv, 7, 2021
|
|
2R7L
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ATP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-TRIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
3CXR
 
 | | Crystal structure of gluconate 5-dehydrogase from streptococcus suis type 2 | | Descriptor: | Dehydrogenase with different specificities | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2008-04-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: Crystal structures of the substrate-free and quaternary complex enzymes.
Protein Sci., 18, 2009
|
|
6PAA
 
 | |
5X8G
 
 | |
2R7N
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ADP and FAICAR | | Descriptor: | 5-(formylamino)-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide, 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
8JV5
 
 | |
6VBY
 
 | | Cinnamate 4-hydroxylase (C4H1) from Sorghum bicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cinnamic acid 4-hydroxylase, GLYCEROL, ... | | Authors: | Zhang, B, Kang, C, Lewis, K.M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor.
Plant Physiol., 183, 2020
|
|
8JV6
 
 | |
6PA5
 
 | |
6PA9
 
 | |
