2WQ6
 
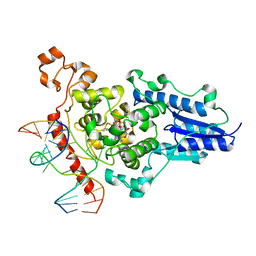 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(Dewar)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*CDWP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions
J.Am.Chem.Soc., 132, 2010
|
|
6DCX
 
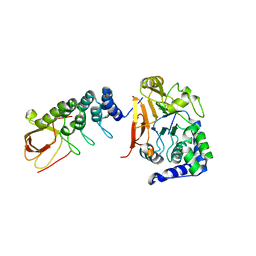 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
6ZEJ
 
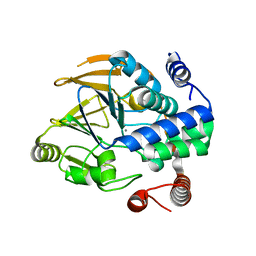 | | Structure of PP1-Phactr1 chimera [PP1(7-304) + linker (SGSGS) + Phactr1(526-580)] | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
2WQ7
 
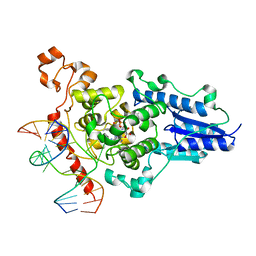 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(6-4)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*ZP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions.
J.Am.Chem.Soc., 132, 2010
|
|
6OBP
 
 | | Reconstituted PP1 holoenzyme | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBN
 
 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4KB8
 
 | |
4KBK
 
 | |
4KBA
 
 | | CK1d in complex with 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine inhibitor | | Descriptor: | 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Liu, S. | | Deposit date: | 2013-04-23 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ligand-protein interactions of selective casein kinase 1 delta inhibitors.
J.Med.Chem., 56, 2013
|
|
4KBC
 
 | |
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
3HVQ
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Neurabin | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Neurabin-1, ... | | Authors: | Critton, D.A, Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4G9J
 
 | | Protein Ser/Thr phosphatase-1 in complex with cell-permeable peptide | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, synthetic peptide | | Authors: | Sukackaite, R, Chatterjee, J, Beullens, M, Bollen, M, Koehn, M, Hart, D.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
8B5R
 
 | | p97-p37-SPI substrate complex | | Descriptor: | I3 sequence being threaded through the p97 channel, Protein phosphatase 1 regulatory subunit 7, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit, ... | | Authors: | van den Boom, J, Marini, G, Meyer, H, Saibil, H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural basis of ubiquitin-independent PP1 complex disassembly by p97.
Embo J., 42, 2023
|
|
3EGG
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Spinophilin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EGH
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1), the PP1 binding and PDZ domains of Spinophilin and the small natural molecular toxin Nodularin-R | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8F2I
 
 | | P53 monomer structure | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Solares, M, Kelly, D.F. | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-Resolution Imaging of Human Cancer Proteins Using Microprocessor Materials.
Chembiochem, 23, 2022
|
|
8F2H
 
 | |
7SD0
 
 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
2WB2
 
 | | Drosophila Melanogaster (6-4) Photolyase Bound To double stranded Dna containing a T(6-4)C Photolesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*64PP*ZP*GP*CP*AP *GP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP*CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Schneider, S, Maul, M.J, Hennecke, U, Carell, T. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the T(6-4)C Lesion in Complex with a (6-4) DNA Photolyase and Repair of Uv- Induced (6-4) and Dewar Photolesions.
Chemistry, 15, 2009
|
|
6LZ3
 
 | |
7XZZ
 
 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7AZT
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AYV
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at cryogenic temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p, ... | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S. | | Deposit date: | 2020-11-13 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3CVY
 
 | | Drosophila melanogaster (6-4) photolyase bound to repaired ds DNA | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanism of a DNA (6-4) photolyase.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
