1UX7
 
 | | Carbohydrate-Binding Module CBM36 in complex with calcium and xylotriose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, SULFATE ION, ... | | Authors: | Davies, G.J, Boraston, A.B, Jamal, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
2GNI
 
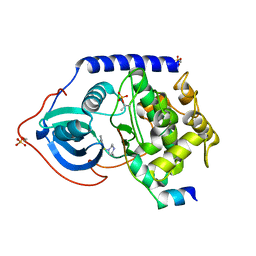 | | PKA fivefold mutant model of Rho-kinase with inhibitor Fasudil (HA1077) | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase inhibitor alpha, cAMP-dependent protein kinase, ... | | Authors: | Bonn, S, Herrero, S, Breitenlechner, C.B, Engh, R.A, Gassel, M, Bossemeyer, D. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
J.Biol.Chem., 281, 2006
|
|
2GMU
 
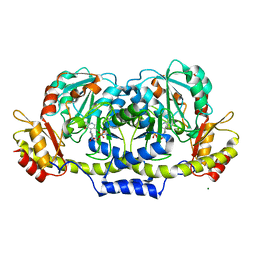 | | Crystal structure of E coli GDP-4-keto-6-deoxy-D-mannose-3-dehydratase complexed with PLP-glutamate ketimine intermediate | | Descriptor: | MAGNESIUM ION, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID, Putative pyridoxamine 5-phosphate-dependent dehydrase, ... | | Authors: | Cook, P.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
Protein Sci., 15, 2006
|
|
1UR3
 
 | |
1LQ0
 
 | |
1LOT
 
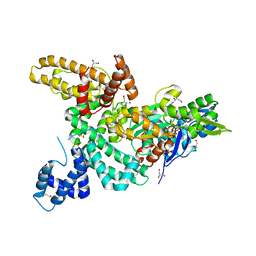 | | CRYSTAL STRUCTURE OF THE COMPLEX OF ACTIN WITH VITAMIN D-BINDING PROTEIN | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Head, J.F, Swamy, N, Ray, R. | | Deposit date: | 2002-05-07 | | Release date: | 2002-07-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex between actin and human vitamin D-binding protein at 2.5 A resolution.
Biochemistry, 41, 2002
|
|
2GSM
 
 | | Catalytic Core (Subunits I and II) of Cytochrome c oxidase from Rhodobacter sphaeroides | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Qin, L, Hiser, C, Mulichak, A, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1LO9
 
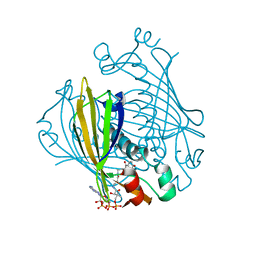 | | X-ray crystal structure of 4-hydroxybenzoyl CoA thioesterase mutant D17N complexed with 4-hydroxybenzoyl CoA | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
2GTQ
 
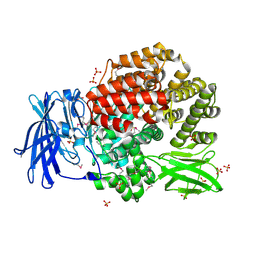 | | Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis | | Descriptor: | SULFATE ION, ZINC ION, aminopeptidase N | | Authors: | Nocek, B, Mulligan, R, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis.
Proteins, 70, 2007
|
|
2GS3
 
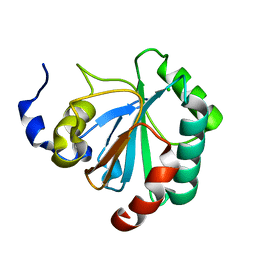 | | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 4(GPX4) | | Descriptor: | CHLORIDE ION, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Johansson, C, Kavanagh, K.L, Rojkova, A, Gileadi, O, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 4(GPX4)
To be Published
|
|
1LRW
 
 | | Crystal structure of methanol dehydrogenase from P. denitrificans | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, methanol dehydrogenase subunit 1, ... | | Authors: | Xia, Z.-X, Dai, W.-W, He, Y.-N, White, S.A, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-05-16 | | Release date: | 2003-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of methanol dehydrogenase from Paracoccus denitrificans and molecular modeling of its interactions with cytochrome c-551i
J.Biol.Inorg.Chem., 8, 2003
|
|
2GSK
 
 | | Structure of the BtuB:TonB Complex | | Descriptor: | CALCIUM ION, CYANOCOBALAMIN, HEXANE, ... | | Authors: | Shultis, D.D, Purdy, M.P, Banchs, C.N, Wiener, M.C. | | Deposit date: | 2006-04-26 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Outer membrane active transport: structure of the BtuB:TonB complex
Science, 312, 2006
|
|
1LT9
 
 | | Crystal Structure of Recombinant Human Fibrinogen Fragment D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Betts, L, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2002-05-20 | | Release date: | 2002-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A Crystal Structures of Recombinant Fibrinogen Fragment D with and without Two Peptide Ligands: GHRP Binding to the "b" Site Disrupts Its Nearby Calcium-binding Site.
Biochemistry, 41, 2002
|
|
1URX
 
 | | Crystallographic structure of beta-agarase A in complex with oligoagarose | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-alpha-D-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose, BETA-AGARASE A, ... | | Authors: | Allouch, J, Helbert, W, Henrissat, B, Czjzek, M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Substrate Binding Sites in a Beta-Agarase Suggest a Novel Mode of Action on Double-Helical Agarose
Structure, 12, 2004
|
|
1NEJ
 
 | | Crystalline Human Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Exhibits The R2 Quaternary State At Neutral pH In The Presence Of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-12-11 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1NKU
 
 | |
1NFW
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR209685 | | Descriptor: | 4-{[(E)-2-(5-CHLOROTHIEN-2-YL)VINYL]SULFONYL}-1-(1H-PYRROLO[3,2-C]PYRIDIN-2-YLMETHYL)PIPERAZIN-2-ONE, CALCIUM ION, COAGULATION FACTOR XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors:
preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
1V9E
 
 | | Crystal Structure Analysis of Bovine Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Saito, R, Sato, T, Ikai, A, Tanaka, N. | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of bovine carbonic anhydrase II at 1.95 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1N9A
 
 | | Farnesyltransferase complex with tetrahydropyridine inhibitors | | Descriptor: | 1-{2-[3-(4-CYANO-BENZYL)-3H-IMIDAZOL-4-YL]-ACETYL}-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: bioavailable analogues with improved cellular potency.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NBX
 
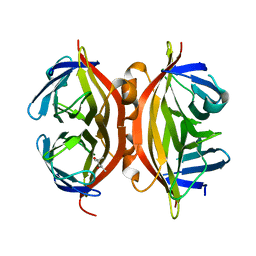 | | Streptavidin Mutant Y43A at 1.70A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1V0J
 
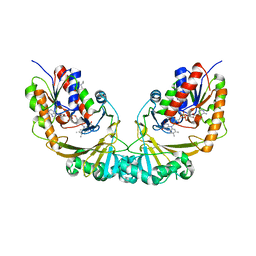 | | Udp-galactopyranose mutase from Mycobacterium tuberculosis | | Descriptor: | BICINE, FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Naismith, J.H. | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state.
J. Mol. Biol., 348, 2005
|
|
1NJB
 
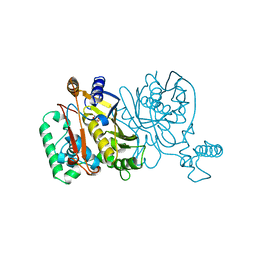 | | THYMIDYLATE SYNTHASE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
1NJA
 
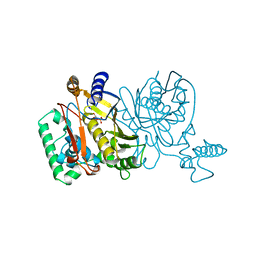 | | THYMIDYLATE SYNTHASE, MUTATION, N229C WITH 2'-DEOXYCYTIDINE 5'-MONOPHOSPHATE (DCMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
1V26
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MYRISTIC ACID, ... | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
1NJD
 
 | | THYMIDYLATE SYNTHASE, MUTATION, N229D WITH 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
