6S0M
 
 | |
7TTO
 
 | |
1M6M
 
 | | V68N MET MYOGLOBIN | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
7TTB
 
 | | P450 (OxyA) from kistamicin biosynthesis, Y99F mutant | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Greule, A, Izore, T, Cryle, M.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801592 Å) | | Cite: | The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade is Highly Sensitive to Oxidative Damage.
Front Chem, 10, 2022
|
|
1CR8
 
 | |
3L6C
 
 | | X-ray crystal structure of rat serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Wood, M, Schonfeld, D, Mather, O, Cesura, A, Barker, J. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
1MNO
 
 | | V68N MYOGLOBIN OXY FORM | | Descriptor: | OXYGEN MOLECULE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
7RNL
 
 | | Yeast CTP Synthase (Ura7) H360R Filament bound to Substrates | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMF
 
 | | Substrate-bound Ura7 filament at low pH | | Descriptor: | CTP synthase | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RL5
 
 | | Yeast CTP Synthase (URA8) filament bound to CTP at low pH | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMO
 
 | | Yeast CTP Synthase (Ura7) Bundle bound to Products at low pH | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RL0
 
 | | Yeast CTP Synthase (URA8) Filament bound to ATP/UTP at low pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-22 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
6XQ8
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 11 | | Descriptor: | 3-phenoxyphenol, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ5
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 1 | | Descriptor: | 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
5K7X
 
 | |
6XQ7
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 5 | | Descriptor: | 5-bromo-3-methyl-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
7MIS
 
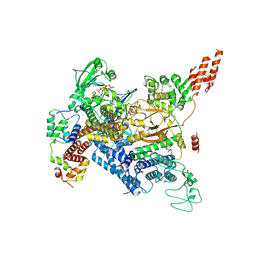 | | Cryo-EM structure of SidJ-SdeC-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
6XQ9
 
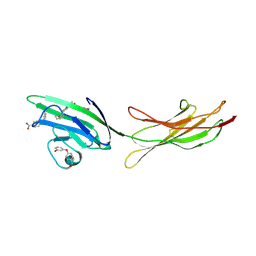 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 13 | | Descriptor: | 4-(3-hydroxyphenoxy)benzoic acid, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
7MIR
 
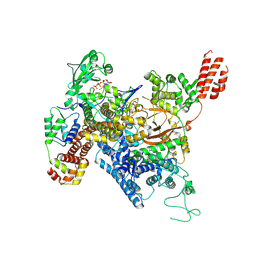 | | Cryo-EM structure of SidJ-SdeA-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
2QWY
 
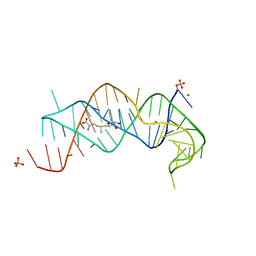 | | SAM-II riboswitch bound to S-adenosylmethionine | | Descriptor: | CESIUM ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gilbert, S.D, Rambo, R.P, Van Tyne, D, Batey, R.T. | | Deposit date: | 2007-08-10 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the SAM-II riboswitch bound to S-adenosylmethionine.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5M96
 
 | |
1MWD
 
 | | WILD TYPE DEOXY MYOGLOBIN | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, A.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
5XPJ
 
 | |
4QY3
 
 | |
8E94
 
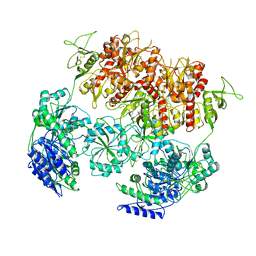 | | PYD-106-bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
