6STP
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with sialic acid | | Descriptor: | Arundo donax Lectin (ADL), GLYCEROL, N-acetyl-alpha-neuraminic acid | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6P7S
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1AXZ
 
 | |
1AY2
 
 | | STRUCTURE OF THE FIBER-FORMING PROTEIN PILIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Parge, H.E, Tainer, J.A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-04-29 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fibre-forming protein pilin at 2.6 A resolution.
Nature, 378, 1995
|
|
6TP2
 
 | |
5WZY
 
 | | Crystal structure of the P2X4 receptor from zebrafish in the presence of CTP at 2.8 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural insights into the nucleotide base specificity of P2X receptors
Sci Rep, 7, 2017
|
|
6TL8
 
 | | Structural basis of SALM3 dimerization and adhesion complex formation with the presynaptic receptor protein tyrosine phosphatases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Myeloid cell surface antigen CD33,Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | | Authors: | Karki, S, Shkumatov, A.V, Bae, S, Ko, J, Kajander, T. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of SALM3 dimerization and synaptic adhesion complex formation with PTP sigma.
Sci Rep, 10, 2020
|
|
6TLZ
 
 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 3'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
4GWW
 
 | |
4GX4
 
 | |
4GX6
 
 | |
3JPY
 
 | | Crystal structure of the zinc-bound amino terminal domain of the NMDA receptor subunit NR2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2009-09-04 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit.
Embo J., 28, 2009
|
|
1W3L
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOTRI DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
6T8K
 
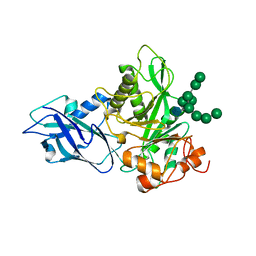 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc product in P1 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
1BYB
 
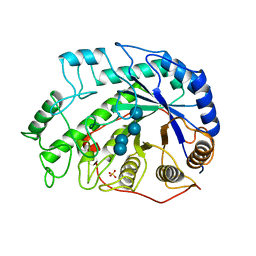 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
6P7Y
 
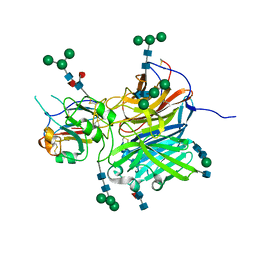 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MAN
 
 | |
1BOS
 
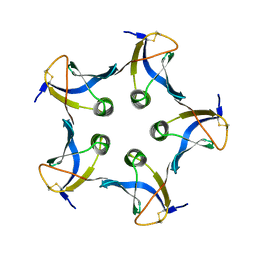 | | SHIGA-LIKE TOXIN COMPLEXED WITH ITS RECEPTOR | | Descriptor: | SHIGA-LIKE TOXIN I B SUBUNIT, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Ling, H, Boodhoo, A, Hazes, B, Cummings, M.D, Armstrong, G.D, Brunton, J.L, Read, R.J. | | Deposit date: | 1998-01-13 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3.
Biochemistry, 37, 1998
|
|
7P8B
 
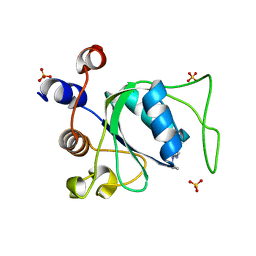 | | Crystal structure of YTHDC1 with compound YLI_DC1_006 | | Descriptor: | 9-cyclopropyl-~{N}-methyl-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6PFO
 
 | |
7P87
 
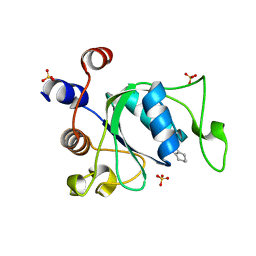 | | Crystal structure of YTHDC1 with compound YLI_DC1_001 | | Descriptor: | N-methyl-4,5,6,7-tetrahydro-1H-indazole-3-carboxamide, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6TCV
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc2Asn substrate | | Descriptor: | ASPARAGINE, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.311 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
1BYC
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
3NKO
 
 | | Crystal structure of mouse autotaxin in complex with 16:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
1BZW
 
 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
