7TE1
 
 | |
5O39
 
 | | Human Brd2(BD2) mutant in complex with ME | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, CHLORIDE ION, ... | | Authors: | Runcie, A.C, Chan, K.-H, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5O3F
 
 | | Human Brd2(BD2) mutant in complex with ET-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-butanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Runcie, A.C, Chan, K.-H, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
4ELD
 
 | |
1O1B
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1XFT
 
 | |
5O6C
 
 | | Crystal Structure of a threonine-selective RCR E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2, ZINC ION | | Authors: | Pao, K.-C, Rafie, K.Z, van Aalten, D, Virdee, S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity.
Nature, 556, 2018
|
|
5IF1
 
 | | Crystal structure apo CDK2/cyclin A | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
6OJR
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 apo-LsdA | | Descriptor: | GLYCEROL, Lignostilbene-alpha,beta-dioxygenase isozyme I, MAGNESIUM ION | | Authors: | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
6HHE
 
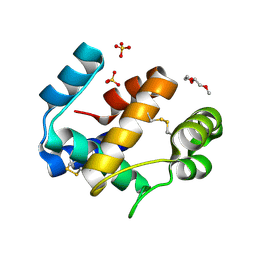 | | Crystal structure of the medfly Odorant Binding Protein CcapOBP22/CcapOBP69a | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Odorant binding protein OBP69a, SULFATE ION | | Authors: | Falchetto, M, Ciossani, G, Nenci, S, Mattevi, A, Gasperi, G, Forneris, F. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication.
Insect Mol.Biol., 28, 2019
|
|
1O1C
 
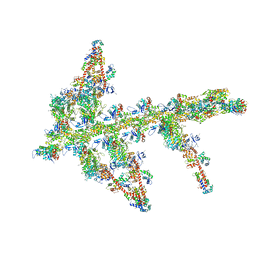 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-18 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
5LIF
 
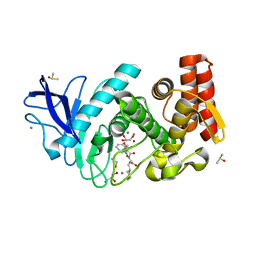 | | Thermolysin in complex with inhibitor | | Descriptor: | (2~{S})-3-cyclohexyl-2-[[(2~{S})-4-methyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]pentanoyl]amino]propanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Elucidating the Origin of Long Residence Time Binding for Inhibitors of the Metalloprotease Thermolysin.
ACS Chem. Biol., 12, 2017
|
|
1XZI
 
 | |
1XZM
 
 | |
1XZG
 
 | |
1XZH
 
 | |
2XPG
 
 | | Crystal structure of a MHC class I-peptide complex | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-3 ALPHA CHAIN, ... | | Authors: | McMahon, R.M, Friis, L, Siebold, C, Friese, M.A, Fugger, L, Jones, E.Y. | | Deposit date: | 2010-08-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Hla-A0301 in Complex with a Peptide of Proteolipid Protein: Insights Into the Role of Hla-A Alleles in Susceptibility to Multiple Sclerosis
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3M2R
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, Coenzyme B, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
1NQ7
 
 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | 7-(3,5-DITERT-BUTYLPHENYL)-3-METHYLOCTA-2,4,6-TRIENOIC ACID, NUCLEAR RECEPTOR ROR-BETA, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
5LCR
 
 | |
3M30
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
4FD9
 
 | |
5ODF
 
 | |
5IEX
 
 | | Crystal structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-hydroxy-1-methylpropyl]oxy}- pyrimidin-2-yl)amino]phenyl}-S-cyclopropylsulfoximide bound to CDK2 | | Descriptor: | (2R,3R)-3-[(5-bromo-2-{[4-(S-cyclopropylsulfonimidoyl)phenyl]amino}pyrimidin-4-yl)oxy]butan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5M5K
 
 | | S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine and cordycepin | | Descriptor: | 3'-DEOXYADENOSINE, ACETATE ION, ADENOSINE, ... | | Authors: | Manszewski, T, Mueller-Dieckamann, J, Jaskolski, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
