1BKT
 
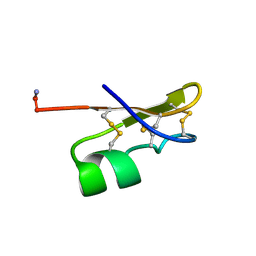 | | BMKTX TOXIN FROM SCORPION BUTHUS MARTENSII KARSCH, NMR, 25 STRUCTURES | | Descriptor: | BMKTX | | Authors: | Renisio, J.G, Romi-Lebrun, R, Blanc, E, Bornet, O, Nakajima, T, Darbon, H. | | Deposit date: | 1998-07-03 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKTX, a K+ blocker toxin from the Chinese scorpion Buthus Martensi
Proteins, 38, 2000
|
|
8OM9
 
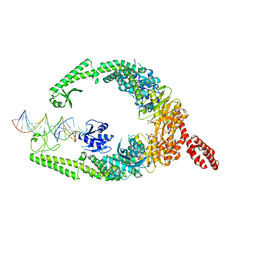 | | MutSbeta bound to (CAG)2 DNA (open form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (open form)
To Be Published
|
|
5KPL
 
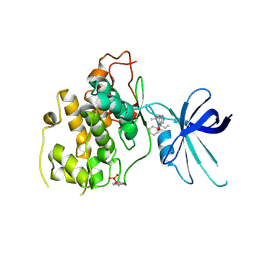 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
1KO0
 
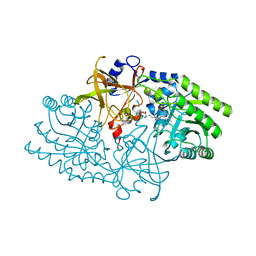 | | Crystal Structure of a D,L-lysine complex of diaminopimelate decarboxylase | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, LYSINE, ... | | Authors: | Levdikov, V, Blagova, L, Bose, N, Momany, C. | | Deposit date: | 2001-12-19 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diaminopimelate Decarboxylase uses a Versatile Active Site for Stereospecific Decarboxylation
To be Published
|
|
3PEG
 
 | | Crystal structure of Neurofibromins Sec14-PH module containing a patient derived duplication (TD) | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, MAGNESIUM ION, Neurofibromin, ... | | Authors: | Welti, S, Kuen, S, D'Angelo, I, Scheffzek, K. | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-08 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.524 Å) | | Cite: | Structural and biochemical consequences of NF1 associated nontruncating mutations in the Sec14-PH module of neurofibromin.
Hum.Mutat., 32, 2011
|
|
1BP1
 
 | | CRYSTAL STRUCTURE OF BPI, THE HUMAN BACTERICIDAL PERMEABILITY-INCREASING PROTEIN | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Beamer, L.J, Carroll, S.F, Eisenberg, D. | | Deposit date: | 1997-04-08 | | Release date: | 1997-09-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution.
Science, 276, 1997
|
|
8OP0
 
 | | VHH Z70 mutant 20 in interaction with PHF6 Tau peptide | | Descriptor: | Tau PHF6 peptide, VHH Z70 mutant 20 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
1KQO
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
8OLX
 
 | | MutSbeta bound to (CAG)2 DNA (canonical form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein Msh2, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (canonical form)
To Be Published
|
|
5ZOE
 
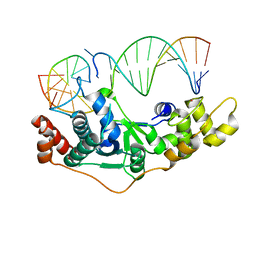 | | Crystal Structure of D181A hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
3PBP
 
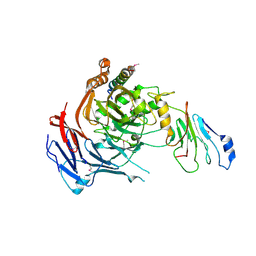 | |
8BQQ
 
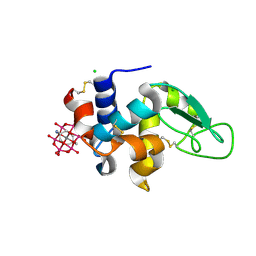 | | Hen Egg-White Lysozyme (HEWL) complexed with amine-functionalised Anderson-Evans polyoxometalate | | Descriptor: | CHLORIDE ION, Lysozyme C, Mn-Mo(6)-N(2)-O(24)-C(8) cluster | | Authors: | Lentink, S, Salazar Marcano, D.E, Moussawi, M.A, Vandebroek, L, Van Meervelt, L, Parac-Vogt, T.N. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Fine-tuning non-covalent interactions between hybrid metal-oxo clusters and proteins.
Faraday Disc.Chem.Soc, 244, 2023
|
|
5DQN
 
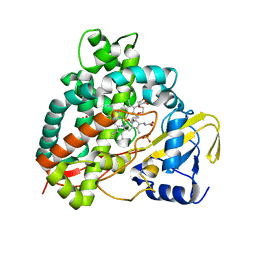 | | Polyethylene 600-bound form of P450 CYP125A3 mutant from Myobacterium Smegmatis - W83Y | | Descriptor: | CITRIC ACID, Cytochrome P450 CYP125, PENTAETHYLENE GLYCOL, ... | | Authors: | Ortiz de Montellano, P.J, Frank, D.J, Waddling, C.A. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Cytochrome P450 125A4, the Third Cholesterol C-26 Hydroxylase from Mycobacterium smegmatis.
Biochemistry, 54, 2015
|
|
8BQR
 
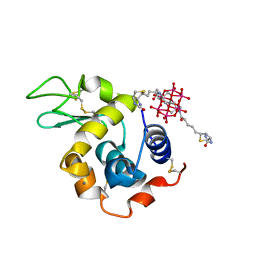 | | Hen Egg-White Lysozyme (HEWL) complexed with biotin-functionalised Anderson-Evans polyoxometalate | | Descriptor: | Anderson-Evans polyoxometalate (biotin-functionalised), CALCIUM ION, Lysozyme C | | Authors: | Lentink, S, Salazar Marcano, D.E, Moussawi, M.A, Vandebroek, L, Van Meervelt, L, Parac-Vogt, T.N. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fine-tuning non-covalent interactions between hybrid metal-oxo clusters and proteins.
Faraday Disc.Chem.Soc, 244, 2023
|
|
8BR9
 
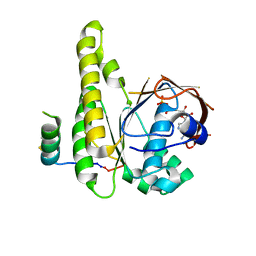 | |
5DJQ
 
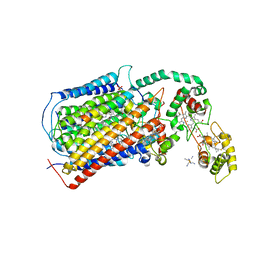 | | The structure of CBB3 cytochrome oxidase. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoN1, ... | | Authors: | Buschmann, S, Warkentin, E, Xie, H, Kohlstaedt, M, Langer, J.D, Ermler, U, Michel, H. | | Deposit date: | 2015-09-02 | | Release date: | 2016-01-13 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
Science, 329, 2010
|
|
3V9I
 
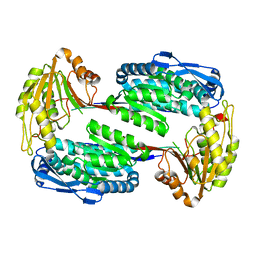 | |
8BBS
 
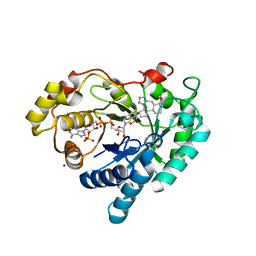 | | Structure of AKR1C3 in complex with a bile acid fused tetrazole inhibitor | | Descriptor: | (4~{R})-4-[(1~{R},2~{S},5~{R},6~{R},13~{S},14~{S},17~{R},19~{R})-6,14-dimethyl-17-oxidanyl-7,8,9,10-tetrazapentacyclo[11.8.0.0^{2,6}.0^{7,11}.0^{14,19}]henicosa-8,10-dien-5-yl]pentanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Petri, E.T, Skerlova, J, Marinovic, M, Brynda, J, Kugler, M, Skoric, D, Bekic, S, Celic, A.S, Rezacova, P. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of human aldo-keto reductase 1C3 in complex with a bile acid fused tetrazole inhibitor: experimental validation, molecular docking and structural analysis.
Rsc Med Chem, 14, 2023
|
|
8OMO
 
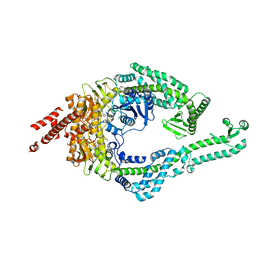 | | DNA-unbound MutSbeta-ATP complex (bent clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (bent clamp form)
To Be Published
|
|
8OM5
 
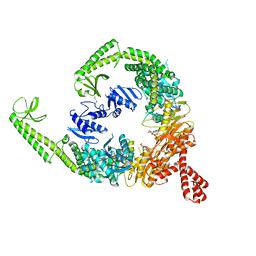 | | DNA-free open form of MutSbeta | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-free open form of MutSbeta
To Be Published
|
|
5VVR
 
 | |
8OMA
 
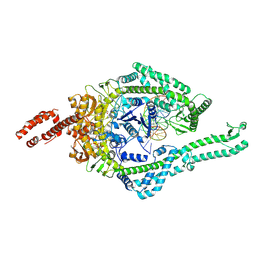 | | MutSbeta bound to 61bp homoduplex DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | MutSbeta bound to 61bp homoduplex DNA
To Be Published
|
|
3PBN
 
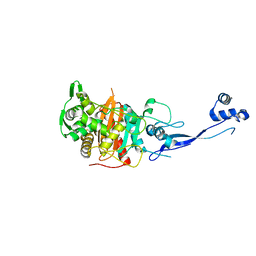 | | Crystal Structure of Apo PBP3 from Pseudomonas aeruginosa | | Descriptor: | Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8OMT
 
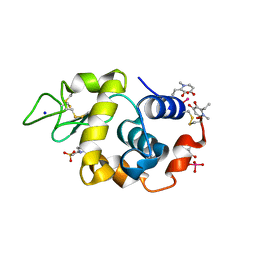 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(empp)2] (Structure C) | | Descriptor: | 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone)V(IV)O4, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2023-03-31 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Implications of Protein Interaction in the Speciation of Potential V IV O-Pyridinone Drugs.
Inorg.Chem., 62, 2023
|
|
3PBS
 
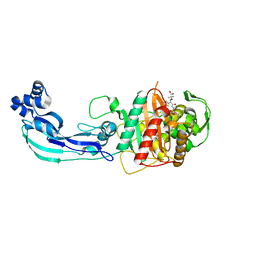 | | Crystal structure of PBP3 complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
