5VIH
 
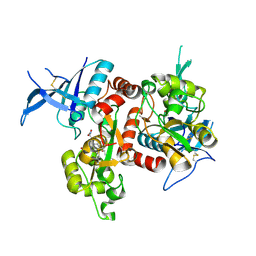 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-fluorophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-fluorophenyl)-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCINE, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WBE
 
 | | Crystal structure of coiled coil region of human septin 1 | | Descriptor: | ACETATE ION, Septin-1, ZINC ION | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
6WBP
 
 | | Crystal structure of coiled coil region of human septin 6 | | Descriptor: | CITRIC ACID, SULFATE ION, Septin-6 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
4OCO
 
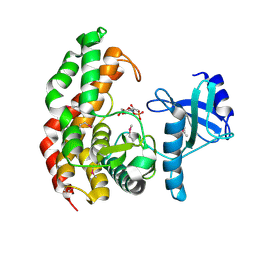 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9FK0
 
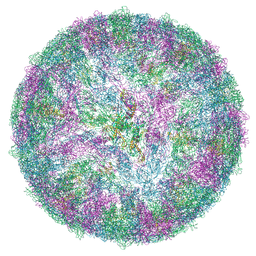 | | LGTV with TBEV prME | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The influence of the pre-membrane and envelope proteins on structure, pathogenicity and tropism of tick-borne encephalitis virus
Biorxiv, 2024
|
|
4OG4
 
 | | Human menin with bound inhibitor MIV-3S | | Descriptor: | 4-(3-{4-[(S)-cyclopentyl(hydroxy)phenylmethyl]piperidin-1-yl}propoxy)benzonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | He, S, Senter, T.J, Pollock, J.W, Han, C, Upadhyay, S.K, Purohit, T, Gogliotti, R.D, Lindsley, C.W, Cierpicki, T, Stauffer, S.R, Grembecka, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Affinity Small-Molecule Inhibitors of the Menin-Mixed Lineage Leukemia (MLL) Interaction Closely Mimic a Natural Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
9JUB
 
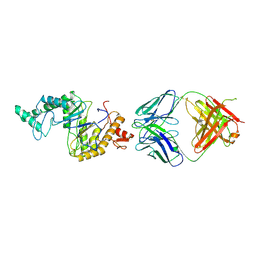 | | Cryo-EM structure of human hyaluronidase PH-20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 79C11Fab, ... | | Authors: | Im, S.-B, Jeong, T.-K, Kim, N, Oh, B.-H. | | Deposit date: | 2024-10-07 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human hyaluronidase PH-20
To Be Published
|
|
5VNG
 
 | |
9DVA
 
 | |
4OH2
 
 | | Crystal Structure of Cu/Zn Superoxide Dismutase I149T | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Crane, B.R, Merz, G.E. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-15 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Copper-Based Pulsed Dipolar ESR Spectroscopy as a Probe of Protein Conformation Linked to Disease States.
Biophys.J., 107, 2014
|
|
5VGA
 
 | | Alternative model for Fab 36-65 | | Descriptor: | Fab 36-65 heavy chain, Fab 36-65 light chain, TRIETHYLENE GLYCOL | | Authors: | Stanfield, R.L, Rupp, B, Wlodawer, A, Dauter, Z, Porebski, P.J, Minor, W, Jaskolski, M, Pozharski, E, Weichenberger, C.X. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6WB4
 
 | | Microbiome-derived Acarbose Kinase Mak1 Labeled with selenomethionine | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Acarbose Kinase Mak1, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
5VGE
 
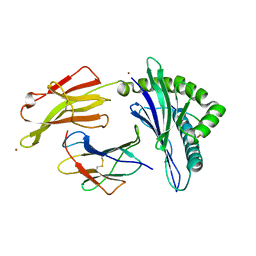 | | Crystal structure of HLA-C*07:02 in complex with RYR peptide | | Descriptor: | ARG-TYR-ARG-PRO-GLY-THR-VAL-ALA-LEU, Beta-2-microglobulin, ZINC ION, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Gras, S, Rossjohn, J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and regulatory diversity shape HLA-C protein expression levels.
Nat Commun, 8, 2017
|
|
4OG3
 
 | | Human menin with bound inhibitor MIV-3R | | Descriptor: | 4-(3-{4-[(R)-cyclopentyl(hydroxy)phenylmethyl]piperidin-1-yl}propoxy)benzonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | He, S, Senter, T.J, Pollock, J.W, Han, C, Upadhyay, S.K, Purohit, T, Gogliotti, R.D, Lindsley, C.W, Cierpicki, T, Stauffer, S.R, Grembecka, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | High-Affinity Small-Molecule Inhibitors of the Menin-Mixed Lineage Leukemia (MLL) Interaction Closely Mimic a Natural Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
9CPB
 
 | |
9DMY
 
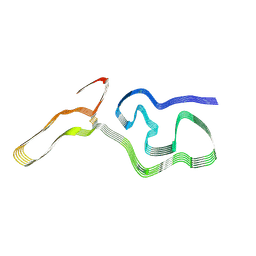 | | Chronic wasting disease prion fibril | | Descriptor: | Major prion protein | | Authors: | Caughey, B, Hoyt, F, Alam, P, Artikis, E, Soukup, J, Hughson, A, Schwartz, C, Race, B, Barbian, K. | | Deposit date: | 2024-09-16 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of a natural prion: chronic wasting disease fibrils from deer.
Acta Neuropathol, 148, 2024
|
|
4OQD
 
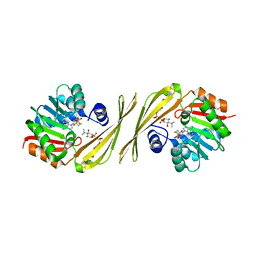 | | Crystal structure of the tylM1 N,N-dimethyltransferase in complex with SAH and TDP-Qui3NMe2 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TDP-3,6-dideoxy-3-N,N-dimethylglucose, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of a novel N-monomethylated dideoxysugar.
Biochemistry, 53, 2014
|
|
9II9
 
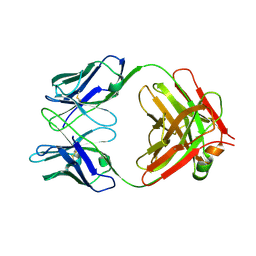 | | Crystal structure of SARS-CoV-2 neutralizing antibody K4-66 | | Descriptor: | antigen-binding fragments (Fabs) | | Authors: | Kimura, T.K, Hashiguchi, T. | | Deposit date: | 2024-06-19 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Induction of IGHV3-53 public antibodies with broadly neutralising activity against SARS-CoV-2 including Omicron subvariants in a Delta breakthrough infection case.
Ebiomedicine, 110, 2024
|
|
5VFP
 
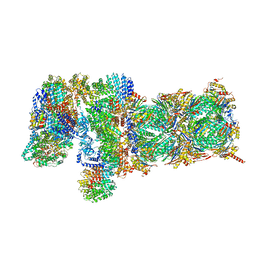 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
6Z0U
 
 | |
9E17
 
 | | Structure of RyR1 in the primed state in the presence of caffeine (reprocessed/reanalyzed from EMPIAR-10997, 7TZC, EMD-26205) | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2024-10-21 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Targeting Ryanodine Receptors with Allopurinol and Xanthine Derivatives for the Treatment of Cardiac and Musculoskeletal Weakness Disorders
To Be Published
|
|
5VJH
 
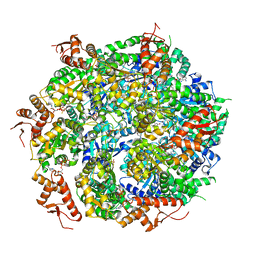 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | Descriptor: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VIF
 
 | | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase | | Descriptor: | 2-{[(2E)-4-chlorobut-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, CKII, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Jiang, J, Li, B, Hu, C.-W, Worth, M, Fan, D, Li, H. | | Deposit date: | 2017-04-15 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase.
Nat. Chem. Biol., 13, 2017
|
|
5VIJ
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-bromophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-bromophenyl)-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6VK9
 
 | | Cryo-EM structure of PilA-N/C from Geobacter sulfurreducens | | Descriptor: | Geopilin domain 1 protein, Geopilin domain 2 protein | | Authors: | Gu, Y, Srikanth, V, Malvankar, N.S, Samatey, F.A. | | Deposit date: | 2020-01-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Geobacter pili reveals secretory rather than nanowire behaviour
Nature, 597, 2021
|
|
