6QES
 
 | | [1-40]Gga-AvBD11 | | Descriptor: | Gallinacin-11 | | Authors: | Meudal, H, Loth, K, Delmas, A.F, Landon, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-12-18 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and evolution ofGga-AvBD11, the archetype of the structural avian-double-beta-defensin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1CGO
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Dobbs, A.J, Faber, H.R, Anderson, B.F, Baker, E.N. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of cytochrome c' from two Alcaligenes species and the implications for four-helix bundle structures.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6K5P
 
 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
6KA1
 
 | | E.coli Malate dehydrogenase | | Descriptor: | Malate dehydrogenase | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
4UH3
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with N1-(3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5- fluorophenyl)-N1,N2-dimethylethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N1-(3-(2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL)-5-FLUOROPHENYL)-N1,N2-DIMETHYLETHANE-1,2-DIAMINE, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | 2-Aminopyridines with a Truncated Side Chain to Improve Human Neuronal Nitric Oxide Synthase Inhibitory Potency and Selectivity.
J.Med.Chem., 58, 2015
|
|
1CP5
 
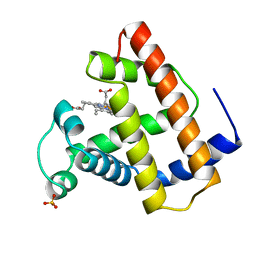 | | RECOMBINANT SPERM WHALE MYOGLOBIN L104F MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
6QG2
 
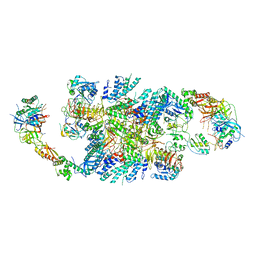 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model A) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QGQ
 
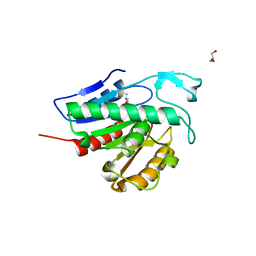 | | Crystal structure of APT1 C2S mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, GLYCEROL, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6KBP
 
 | | Crystal structure of human D-amino acid oxidase mutant (P219L) complexed with benzoate | | Descriptor: | ACETATE ION, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Rachadech, W, Kato, Y, Maita, N, Fukui, K. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human D-amino acid oxidase mutant (P219L) complexed with benzoate
To Be Published
|
|
6QAO
 
 | |
6KCO
 
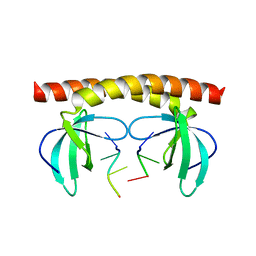 | | Shuguo PWWP in complex with ssDNA | | Descriptor: | DNA (5'-D(*TP*CP*CP*CP*T)-3'), LD23804p | | Authors: | Liu, Y.C, Huang, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shuguo PWWP in complex with ssDNA
To Be Published
|
|
6KD9
 
 | | Binding pose 1 of 2-CF3 bound AsqJ complex | | Descriptor: | (3~{Z})-4-methyl-3-[[4-(trifluoromethyl)phenyl]methylidene]-1~{H}-1,4-benzodiazepine-2,5-dione, FE (III) ION, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, ... | | Authors: | Liao, H.J, Chan, N.L. | | Deposit date: | 2019-07-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Binding pose 1 of 2-CF3 bound AsqJ complex
J.Am.Chem.Soc., 2020
|
|
6KDL
 
 | | Crystal structure of human DNMT3B-DNMT3L complex (I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.274 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
6KDY
 
 | |
1CMJ
 
 | |
1CRI
 
 | |
6KCF
 
 | |
6KJ7
 
 | | E. coli ATCase catalytic subunit mutant - G166P | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6Q2U
 
 | | Structure of Cytochrome C4 from Pseudomonas aeruginosa | | Descriptor: | Cytochrome c4, HEME C | | Authors: | Carpenter, J.M, Zhong, F, Pletneva, E.V, Ragusa, M.J. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and redox properties of the diheme electron carrier cytochrome c4from Pseudomonas aeruginosa.
J.Inorg.Biochem., 203, 2019
|
|
6KJM
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
6KJT
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | Putative beta-lactamase, SUCCINIC ACID | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
4UHA
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5-(methyl(2-(methylamino)ethyl)amino)benzonitrile | | Descriptor: | 3-(2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL)-5-(METHYL(2-(METHYLAMINO)ETHYL)AMINO)BENZONITRILE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Aminopyridines with a Truncated Side Chain To Improve Human Neuronal Nitric Oxide Synthase Inhibitory Potency and Selectivity.
J. Med. Chem., 58, 2015
|
|
4UR3
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans P2(1) crystal form | | Descriptor: | IRON/SULFUR CLUSTER, NORPSEUDO-B12, TETRACHLOROETHENE REDUCTIVE DEHALOGENASE CATALYTIC SUBUNIT | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
6KEZ
 
 | | Crystal structure of GAPDH/CP12/PRK complex from Arabidopsis thaliana | | Descriptor: | Calvin cycle protein CP12-2, Glyceraldehyde-3-phosphate dehydrogenase GAPA1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
6KGF
 
 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 8.2 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
