8XLJ
 
 | |
6MTI
 
 | | Synaptotagmin-1 C2A, C2B domains and SNARE-pin proteins (5CCI) individually docked into Cryo-EM map of C2AB-SNARE complexes helically organized on lipid nanotube surface in presence of Mg2+ | | Descriptor: | MAGNESIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Grushin, K, Wang, J, Coleman, J, Rothman, J, Sindelar, C, Krishnakumar, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural basis for the clamping and Ca2+activation of SNARE-mediated fusion by synaptotagmin.
Nat Commun, 10, 2019
|
|
6Y8C
 
 | |
3U60
 
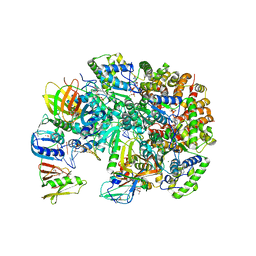 | | Structure of T4 Bacteriophage Clamp Loader Bound To Open Clamp, DNA and ATP Analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
7YQP
 
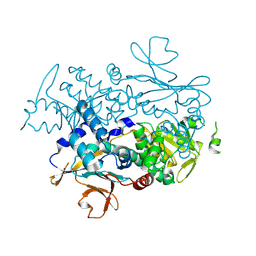 | | Xcc Nicotinamide Phosphoribosyltransferase | | Descriptor: | Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7YQO
 
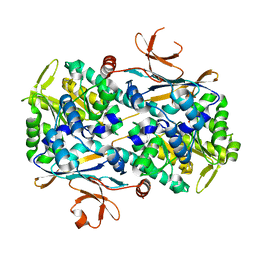 | | Xcc Nicotinamide Phosphoribosyltransferase | | Descriptor: | GLYCEROL, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
5FRQ
 
 | |
8FU6
 
 | | GCGR-Gs complex in the presence of RAMP2 | | Descriptor: | Glucagon derivative ZP3780, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, O'Brien, E.S, Wang, H, Montabana, E, Kobilka, B.K. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Negative allosteric modulation of the glucagon receptor by RAMP2.
Cell, 186, 2023
|
|
4YS0
 
 | |
8Y57
 
 | | Anti MDMA antibody 1bB11 complex | | Descriptor: | (2S)-1-(1,3-benzodioxol-5-yl)-N-methylpropan-2-amine, Heavy chain of 1bB11, Light chain of 1bB11 | | Authors: | Cheon, G, Lee, Y, Lee, S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an antibody specifically recognizing 3,4-methyl enedioxy methamphetamine through the epoxide moiety.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
1UNN
 
 | |
5G4Q
 
 | | H.pylori Beta clamp in complex with 5-chloroisatin | | Descriptor: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
5G48
 
 | | H.pylori Beta clamp in complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting the beta-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal.
FEBS Lett., 591, 2017
|
|
3U5Z
 
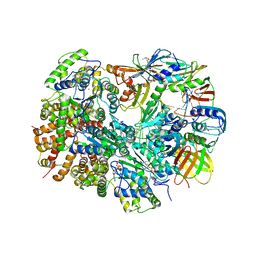 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
3U61
 
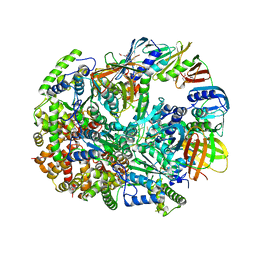 | | Structure of T4 Bacteriophage Clamp Loader Bound To Closed Clamp, DNA and ATP Analog and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
2KEC
 
 | |
2KED
 
 | |
8FQ1
 
 | |
8FQ5
 
 | |
8FP4
 
 | |
8FP9
 
 | | GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FQB
 
 | | GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma2, with 10mM CaCl2, 140mM NMDG, 330uM CTZ, and 100mM L-glutamate (Open-Ca10) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPG
 
 | | GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100uM CNQX (Closed-CaNaMg) | | Descriptor: | CHLORIDE ION, Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPV
 
 | |
8FPY
 
 | |
