6FE8
 
 | | Cryo-EM structure of the core Centromere Binding Factor 3 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Zhang, W.J, Lukoynova, N, Miah, S, Vaughan, C.K. | | Deposit date: | 2017-12-30 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
4PHT
 
 | | ATPase GspE in complex with the cytoplasmic domain of GspL from the Vibrio vulnificus type II Secretion system | | Descriptor: | General secretory pathway protein E, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lu, C, Korotkov, K, Hol, W. | | Deposit date: | 2014-05-06 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of the full-length ATPase GspE from the Vibrio vulnificus type II secretion system in complex with the cytoplasmic domain of GspL.
J.Struct.Biol., 187, 2014
|
|
6FO1
 
 | | Human R2TP subcomplex containing 1 RUVBL1-RUVBL2 hexamer bound to 1 RBD domain from RPAP3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA polymerase II-associated protein 3, RuvB-like 1, ... | | Authors: | Martino, F, Munoz-Hernandez, H, Rodriguez, C.F, Pearl, L.H, Llorca, O. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-04 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex.
Nat Commun, 9, 2018
|
|
5A42
 
 | | Cryo-EM single particle 3D reconstruction of the native conformation of E. coli alpha-2-macroglobulin (ECAM) | | Descriptor: | UNCHARACTERIZED LIPOPROTEIN YFHM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, F.X. | | Deposit date: | 2015-06-04 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural and Functional Insights Into Escherichia Coli Alpha2- Macroglobulin Endopeptidase Snap-Trap Inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A3S
 
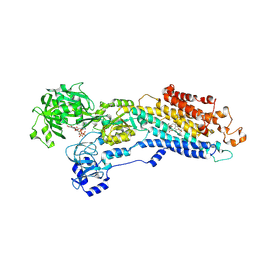 | | Crystal structure of the (SR) Calcium ATPase E2-vanadate complex bound to thapsigargin and TNP-ATP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
5EHM
 
 | |
4RZ7
 
 | | Crystal Structure of PVX_084705 with bound PCI32765 | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, UNKNOWN ATOM OR ION, cGMP-dependent protein kinase, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal Structure of PVX_084705 with bound PCI32765
To be Published
|
|
6G2T
 
 | | human cardiac myosin binding protein C C1 Ig-domain bound to native cardiac thin filament | | Descriptor: | Actin, cytoplasmic 2, Myosin-binding protein C, ... | | Authors: | Risi, C, Belknap, B, Forgacs, E, Harris, S.P, Schroder, G.F, White, H.D, Galkin, V.E. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | N-Terminal Domains of Cardiac Myosin Binding Protein C Cooperatively Activate the Thin Filament.
Structure, 26, 2018
|
|
4A3B
 
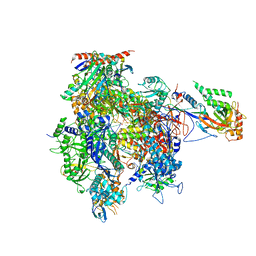 | | RNA Polymerase II initial transcribing complex with a 4nt DNA-RNA hybrid | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
1W46
 
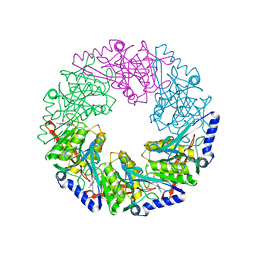 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
6C0A
 
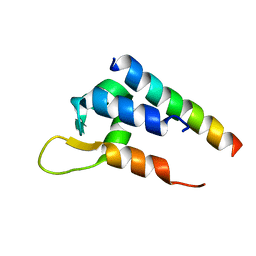 | | Actinin-1 EF-Hand bound to the Cav1.2 IQ Motif | | Descriptor: | Alpha-actinin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Turner, M.L, Ames, J.B. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Localization and Activation of Neuronal L-type Ca2+ Channels by a-Actinin1 and Calmodulin
To be Published
|
|
1W47
 
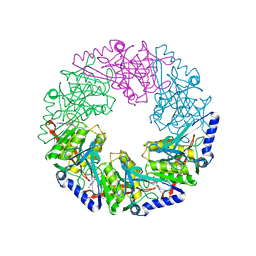 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
5EHS
 
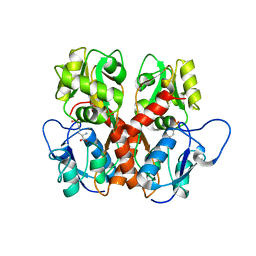 | |
4A93
 
 | | RNA Polymerase II elongation complex containing a CPD Lesion | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*T*TTP*TP*TP*CP*C BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP*AP)-3', ... | | Authors: | Walmacq, C, Cheung, A.C.M, Kireeva, M.L, Lubkowska, L, Ye, C, Gotte, D, Strathern, J.N, Carell, T, Cramer, P, Kashlev, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Translesion Transcription by RNA Polymerase II and its Role in Cellular Resistance to DNA Damage.
Mol.Cell, 46, 2012
|
|
6DMW
 
 | |
6DNF
 
 | |
6C0F
 
 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | Descriptor: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2017-12-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
6BJO
 
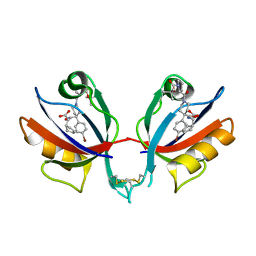 | | PICK1 PDZ domain in complex with the small molecule inhibitor BIO124. | | Descriptor: | (2S)-({4-(4-bromophenyl)-1-[1-(tert-butoxycarbonyl)-L-prolyl]piperidine-4-carbonyl}amino)(cyclopentyl)acetic acid, PRKCA-binding protein | | Authors: | Marcotte, D.J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Lock and chop: A novel method for the generation of a PICK1 PDZ domain and piperidine-based inhibitor co-crystal structure.
Protein Sci., 27, 2018
|
|
6BJN
 
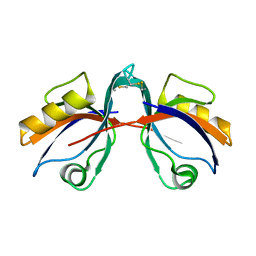 | |
4RBS
 
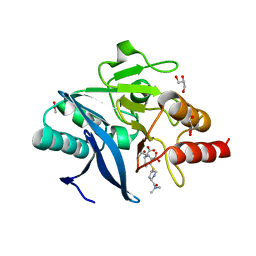 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, ACETIC ACID, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem
To be Published
|
|
5FX2
 
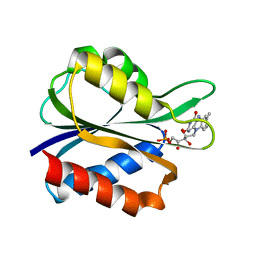 | |
6G0L
 
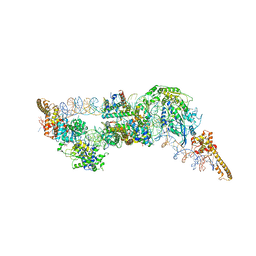 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
5FET
 
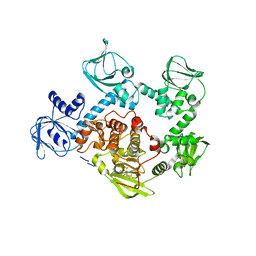 | | Crystal Structure of PVX_084705 in presence of Compound 2 | | Descriptor: | 4-[7-[(dimethylamino)methyl]-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl]pyrimidin-2-amine, cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, Walker, J.R, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of PVX_084705 in presence of Compound 2
To be published
|
|
6B0I
 
 | | Apo KLP10A in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
6DBO
 
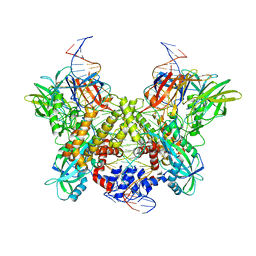 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
