8XJ3
 
 | |
9F9A
 
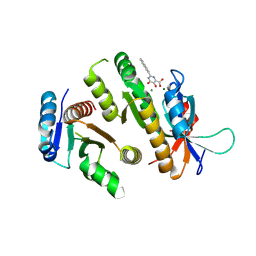 | | Crystal structure of MUS81-EME1 bound by compound 12. | | Descriptor: | 2-naphthalen-2-yl-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9G5Z
 
 | |
8YB5
 
 | |
8ZAK
 
 | |
8ZX6
 
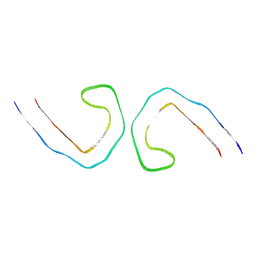 | | Heparin bound Tau fibril PHF | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Tao, Y.Q, Liu, C, Li, D. | | Deposit date: | 2024-06-13 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Heparin bound Tau fibril PHF
To Be Published
|
|
9BRZ
 
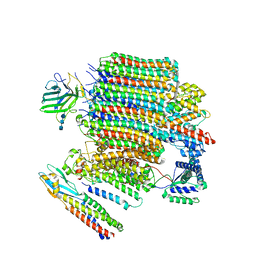 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
8ZNC
 
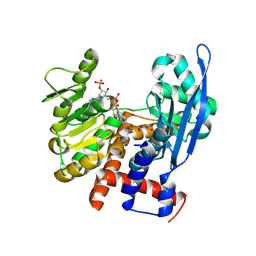 | |
1EQN
 
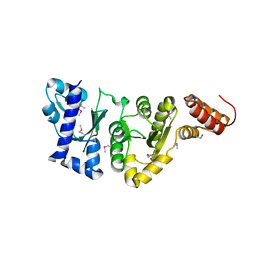 | | E.COLI PRIMASE CATALYTIC CORE | | Descriptor: | DNA PRIMASE | | Authors: | Podobnik, M, McInerney, P, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-04-05 | | Release date: | 2000-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases.
J.Mol.Biol., 300, 2000
|
|
1EUA
 
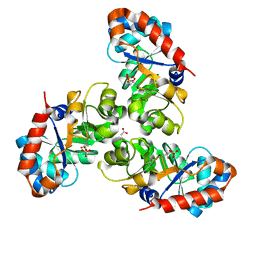 | | SCHIFF BASE INTERMEDIATE IN KDPG ALDOLASE FROM ESCHERICHIA COLI | | Descriptor: | ACETATE ION, KDPG ALDOLASE, PYRUVIC ACID, ... | | Authors: | Allard, J, Grochulski, P, Sygusch, J. | | Deposit date: | 2000-04-14 | | Release date: | 2001-02-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1EUC
 
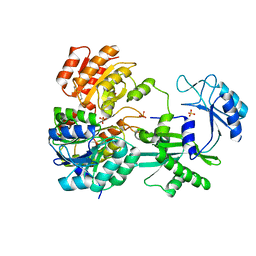 | | CRYSTAL STRUCTURE OF DEPHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE | | Descriptor: | PHOSPHATE ION, SUCCINYL-COA SYNTHETASE, ALPHA CHAIN, ... | | Authors: | Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 2000-04-14 | | Release date: | 2000-07-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase.
J.Mol.Biol., 299, 2000
|
|
1EC9
 
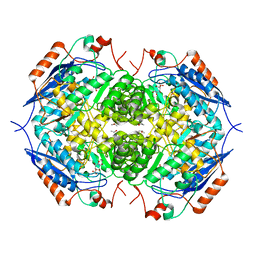 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO XYLAROHYDROXAMATE | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EEZ
 
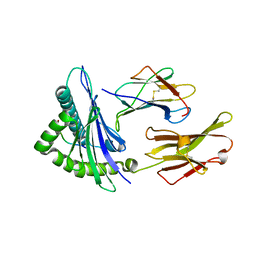 | |
1EI7
 
 | |
1EL2
 
 | |
1EVG
 
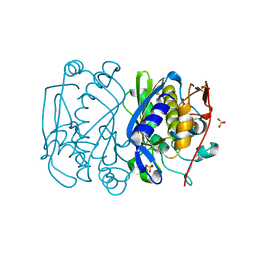 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI WITH UNMODIFIED CATALYTIC CYSTEINE | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
1EV2
 
 | | CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 2), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 2), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EGD
 
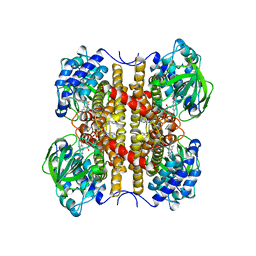 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EGC
 
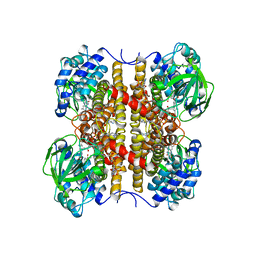 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE COMPLEXED WITH OCTANOYL-COA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE, OCTANOYL-COENZYME A | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
1EFN
 
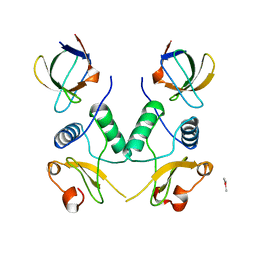 | |
1ERD
 
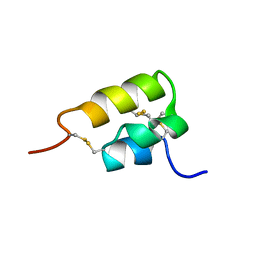 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-2 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-2 | | Authors: | Ottiger, M, Szyperski, T, Luginbuhl, P, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
1ECG
 
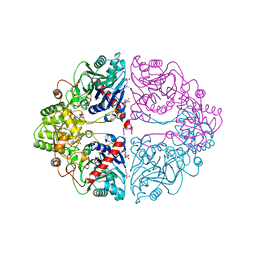 | | DON INACTIVATED ESCHERICHIA COLI GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE (PRPP) AMIDOTRANSFERASE | | Descriptor: | 5-OXO-L-NORLEUCINE, GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE AMIDOTRANSFERASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Krahn, J.M. | | Deposit date: | 1996-04-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site.
J.Biol.Chem., 271, 1996
|
|
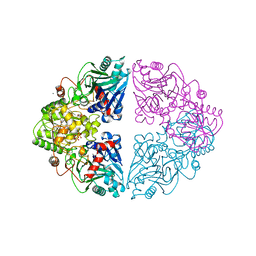
1ECC
 
 | |
1ECB
 
 | |
