6GVN
 
 | | Tubulin:TM-3 DARPin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Cantos Fernandes, S, Campanacci, V. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GVM
 
 | | Tubulin:F3II DARPin complex | | Descriptor: | F3II DARPIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V, Cantos Fernandes, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EE8
 
 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (60-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
8YZ7
 
 | |
2MJ2
 
 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
5IER
 
 | |
5IF6
 
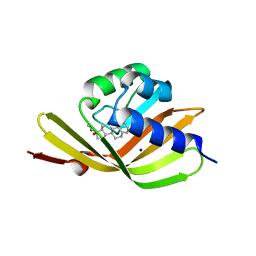 | |
8ZGZ
 
 | |
8XLK
 
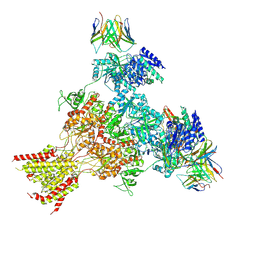 | | Structure of native tri-heteromeric GluN1-GluN2A-GluN2B NMDA receptor in rat cortex and hippocampus | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, M, Feng, J, Li, Y, Zhu, S. | | Deposit date: | 2023-12-26 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Assembly and architecture of endogenous NMDA receptors in adult cerebral cortex and hippocampus.
Cell, 188, 2025
|
|
8W2Z
 
 | | Cas9d 6bp R-loop Seed Complex | | Descriptor: | DNA Non-Target Strand, DNA Target Strand, HNH nuclease domain-containing protein, ... | | Authors: | Fregoso Ocampo, R, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | DNA targeting by compact Cas9d and its resurrected ancestor.
Nat Commun, 16, 2025
|
|
8W2S
 
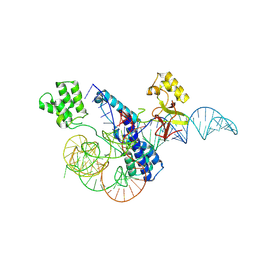 | |
8FA5
 
 | |
2KE5
 
 | | Solution structure and dynamics of the small GTPase Ralb in its active conformation: significance for effector protein binding | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-B | | Authors: | Fenwick, R, Prasannan, S, Campbell, L.J, Nietlispach, D, Evetts, K.A, Camonis, J, Mott, H.R, Owen, D. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the small GTPase RalB in its active conformation: significance for effector protein binding
Biochemistry, 48, 2009
|
|
2J4I
 
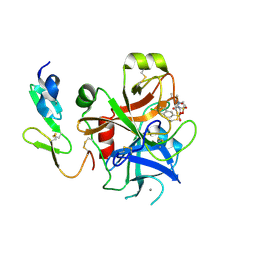 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 1-PYRROLIDINEACETAMIDE, 3-[[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]AMINO]-ALPHA-METHYL-N-(1-METHYLETHYL)-N-[2-[(METHYLSULFONYL)AMINO]ETHYL]-2-OXO-, (ALPHAS,3S)-, ... | | Authors: | Young, R.J, Campbell, M, Borthwick, A.D, Brown, D, Chan, C, Convery, M.A, Crowe, M.C, Dayal, S, Diallo, H, Kelly, H.A, Paul King, N, Kleanthous, S, Kurtis, C.L, Mason, A.M, Mordaunt, J.E, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Smith, P.W, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure- and Property-Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Acyclic Alanyl Amides as P4 Motifs.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4YB0
 
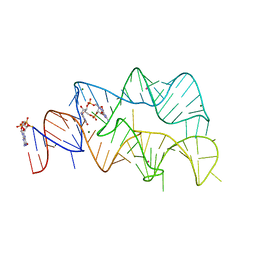 | | 3',3'-cGAMP riboswitch bound with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
2K42
 
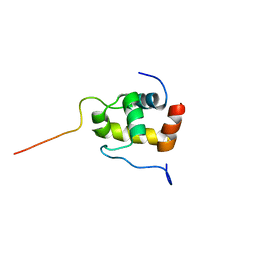 | | Solution Structure of the GTPase Binding Domain of WASP in Complex with EspFU, an EHEC Effector | | Descriptor: | ESPFU, Wiskott-Aldrich syndrome protein | | Authors: | Cheng, H.-C, Skehan, B.M, Campellone, K.G, Leong, J.M, Rosen, M.K. | | Deposit date: | 2008-05-27 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U).
Nature, 454, 2008
|
|
6NJN
 
 | |
6GWC
 
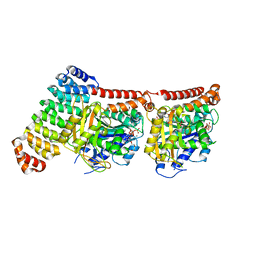 | | Tubulin:iE5 alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALPHA-TUBULIN, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-22 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Structure, 27, 2019
|
|
6GWD
 
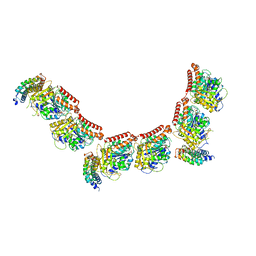 | | Tubulin:iiH5 alphaRep complex | | Descriptor: | ALPHA-TUBULIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-22 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Structure, 27, 2019
|
|
6NJL
 
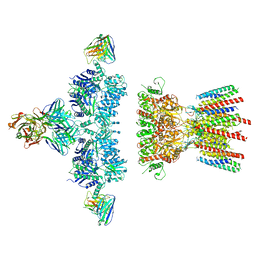 | |
9EUO
 
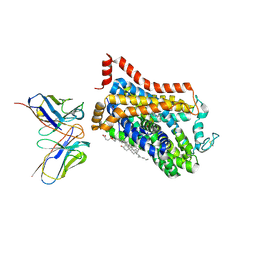 | | Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 168, 2024
|
|
9EUP
 
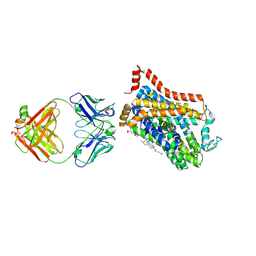 | | Inhibitor-free outward-open structure of Drosophila dopamine transporter | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 168, 2024
|
|
2KN8
 
 | | NMR structure of the C-terminal domain of pUL89 | | Descriptor: | DNA cleavage and packaging protein large subunit, UL89 | | Authors: | Couvreux, A, Hantz, S, Marquant, R, Champier, G, Alain, S, Morellet, N, Bouaziz, S. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the structure of the pUL89 C-terminal domain of the human cytomegalovirus terminase complex.
Proteins, 78, 2010
|
|
7NF9
 
 | | N-terminal C2H2 Zn-finger domain of Clamp | | Descriptor: | Chromatin-linked adaptor for MSL proteins, isoform A, ZINC ION | | Authors: | Mariasina, S.S, Bonchuk, A.N, Tikhonova, E.A, Efimov, S.V, Maksimenko, O.G, Georgiev, P.G, Polshakov, V.I. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for interaction between CLAMP and MSL2 proteins involved in the specific recruitment of the dosage compensation complex in Drosophila.
Nucleic Acids Res., 50, 2022
|
|
8XLL
 
 | | Structure of the native 2-oxoglutarate dehydrogenase complex (OGDHC) in the adult cortex and hippocampus | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Zhang, M, Feng, J, Li, Y, Zhu, S. | | Deposit date: | 2023-12-26 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Assembly and architecture of endogenous NMDA receptors in adult cerebral cortex and hippocampus.
Cell, 188, 2025
|
|
