3JQ4
 
 | | The structure of the complex of the large ribosomal subunit from D. Radiodurans with the antibiotic lankacidin | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, N-[(1S,2R,3E,5E,7S,9E,11E,13S,15R,19R)-7,13-dihydroxy-1,4,10,19-tetramethyl-17,18-dioxo-16-oxabicyclo[13.2.2]nonadeca-3,5,9,11-tetraen-2-yl]-2-oxopropanamide | | Authors: | Auerbach-Nevo, T, Mermershtain, I, Davidovich, C, Bashan, A, Rozenberg, H, Yonath, A. | | Deposit date: | 2009-09-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1K8G
 
 | |
3AU5
 
 | |
3RVD
 
 | | Crystal structure of the binary complex, obtained by soaking, of photosyntetic a4 glyceraldehyde 3-phosphate dehydrogenase (gapdh) with cp12-2, both from arabidopsis thaliana. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Thumiger, A, Falini, G, Marri, L, Sparla, F, Trost, P. | | Deposit date: | 2011-05-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Selection and Folding-upon-binding of Intrinsically Disordered Protein CP12 Regulate Photosynthetic Enzymes Assembly.
J.Biol.Chem., 287, 2012
|
|
3AU4
 
 | |
1Q89
 
 | |
1Q88
 
 | |
1Q87
 
 | |
2FCC
 
 | | Crystal Structure of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed with a DNA Substrate Containing Abasic Site | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3'), Endonuclease V, ... | | Authors: | Golan, G, Zharkov, D.O, Fernandes, A.S, Dodson, M.L, McCullough, A.K, Grollman, A.P, Lloyd, R.S, Shoham, G. | | Deposit date: | 2005-12-12 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of T4 Pyrimidine Dimer Glycosylase in a Reduced Imine Covalent Complex with Abasic Site-containing DNA.
J.Mol.Biol., 362, 2006
|
|
3GDF
 
 | | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum. | | Descriptor: | Probable NADP-dependent mannitol dehydrogenase, ZINC ION | | Authors: | Nuess, D, Goettig, P, Magler, I, Denk, U, Breitenbach, M, Schneider, P.B, Brandstetter, H, Simon-Nobbe, B. | | Deposit date: | 2009-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum: Implications for oligomerisation and catalysis.
Biochimie, 92, 2010
|
|
5J43
 
 | |
2QKM
 
 | |
6WRZ
 
 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 with 7-methyl-GpppA and S-adenosyl-L-homocysteine in the Active Site and Sulfates in the mRNA Binding Groove. | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
4ENC
 
 | | Crystal structure of fluoride riboswitch | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
4FXE
 
 | |
5TWJ
 
 | | Crystal Structure of RlmH in Complex with S-Adenosylmethionine | | Descriptor: | Ribosomal RNA large subunit methyltransferase H, S-ADENOSYLMETHIONINE | | Authors: | Koh, C.S, Madireddy, R, Beane, T.J, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Small methyltransferase RlmH assembles a composite active site to methylate a ribosomal pseudouridine.
Sci Rep, 7, 2017
|
|
3IQR
 
 | |
3EAR
 
 | |
3IQN
 
 | |
3IQP
 
 | |
1UET
 
 | |
3EAS
 
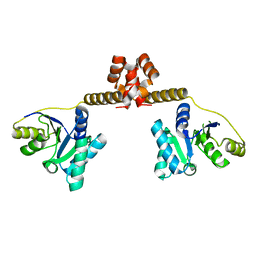 | |
3IYO
 
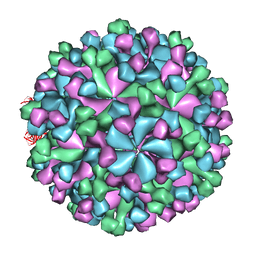 | | Cryo-EM model of virion-sized HEV virion-sized capsid | | Descriptor: | Capsid protein | | Authors: | Xing, L, Mayazaki, N, Li, T.C, Simons, M.N, Wall, J.S, Moore, M, Wang, C.Y, Takeda, N, Wakita, T, Miyamura, T, Cheng, R.H. | | Deposit date: | 2010-03-19 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Structural basis for the RNA-dependent assembly pathway of hepatitis E virion-sized particles
J.Biol.Chem., 2010
|
|
2YIF
 
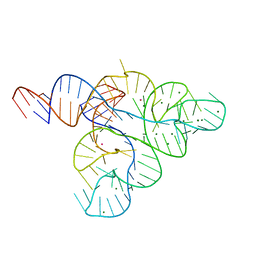 | |
3GDG
 
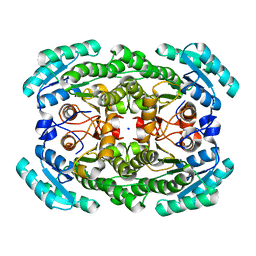 | | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum. | | Descriptor: | Probable NADP-dependent mannitol dehydrogenase, SODIUM ION | | Authors: | Nuess, D, Goettig, P, Magler, I, Denk, U, Breitenbach, M, Schneider, P.B, Brandstetter, H, Simon-Nobbe, B. | | Deposit date: | 2009-02-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum: Implications for oligomerisation and catalysis.
Biochimie, 92, 2010
|
|
