6SDF
 
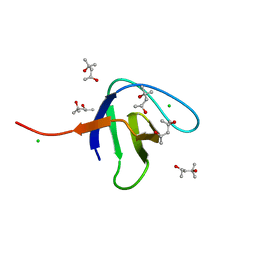 | | N-terminal SH3 domain of Grb2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Growth factor receptor-bound protein 2 | | Authors: | Bolgov, A.A, Korban, S.A, Luzik, D.A, Rogacheva, O.N, Zhemkov, V.A, Kim, M, Skrynnikov, N.R, Bezprozvanny, I.B. | | Deposit date: | 2019-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the SH3 domain of growth factor receptor-bound protein 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8U3Y
 
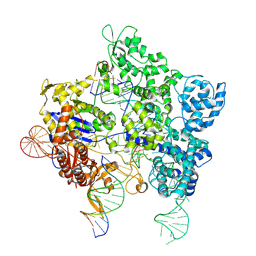 | | SpG Cas9 with NGG PAM DNA target | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
7OL1
 
 | |
8W7P
 
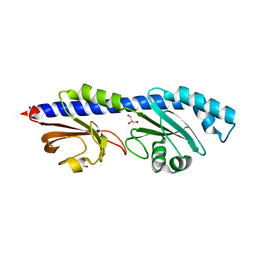 | | Extracellular domain of a sensor histidine kinase | | Descriptor: | Extracellular domain of a sensor histidine kinase NagS, GLYCEROL | | Authors: | Itoh, T, Ogawa, T, Hibi, T, Kimoto, H. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of the extracellular domain of sensor histidine kinase NagS from Paenibacillus sp. str. FPU-7: nagS interacts with oligosaccharide binding protein NagB1 in complexes with N, N'-diacetylchitobiose.
Biosci.Biotechnol.Biochem., 88, 2024
|
|
7BT2
 
 | | Crystal structure of the SERCA2a in the E2.ATP state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kabashima, Y, Ogawa, H, Nakajima, R, Toyoshima, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.00002861 Å) | | Cite: | What ATP binding does to the Ca2+pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7OPN
 
 | | Human Aldehyde Oxidase SNP R1231H in complex with Raloxifene | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Aldehyde oxidase, DIMETHYL SULFOXIDE, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
7ORC
 
 | | Human Aldehyde Oxidase in complex with Raloxifene | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-05 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
8WM0
 
 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
7RAC
 
 | |
7R1O
 
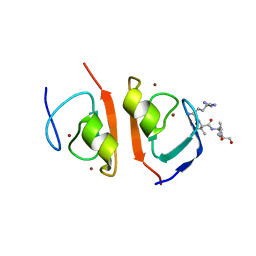 | | p62-ZZ domain of the human sequestosome in complex with dusquetide | | Descriptor: | Dusquetide, Sequestosome-1, ZINC ION | | Authors: | Hakansson, M, Hansson, M, Logan, D.T, Rozek, A, Donini, O. | | Deposit date: | 2022-02-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Dusquetide modulates innate immune response through binding to p62.
Structure, 30, 2022
|
|
7AUA
 
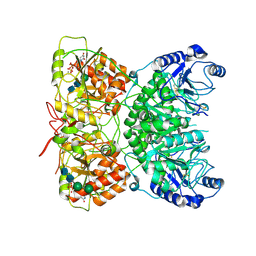 | | Cryo-EM structure of human exostosin-like 3 (EXTL3) in complex with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, MANGANESE (II) ION, ... | | Authors: | Wilson, L.F.L, Dendooven, T, Hardwick, S.W, Chirgadze, D.Y, Luisi, B.F, Logan, D.T, Mani, K, Dupree, P. | | Deposit date: | 2020-11-02 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.
Nat Commun, 13, 2022
|
|
7AU2
 
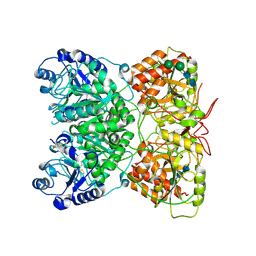 | | Cryo-EM structure of human exostosin-like 3 (EXTL3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wilson, L.F.L, Dendooven, T, Hardwick, S.W, Chirgadze, D.Y, Luisi, B.F, Logan, D.T, Mani, K, Dupree, P. | | Deposit date: | 2020-11-02 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.
Nat Commun, 13, 2022
|
|
6T2T
 
 | | Crystal structure of Drosophila melanogaster glutathione S-transferase epsilon 14 in complex with glutathione and 2-methyl-2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Skerlova, J, Lindstrom, H, Sjodin, B, Gonis, E, Neiers, F, Mannervik, B, Stenmark, P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and steroid isomerase activity of Drosophila glutathione transferase E14 essential for ecdysteroid biosynthesis.
Febs Lett., 594, 2020
|
|
7RRN
 
 | |
6S7Y
 
 | |
7B8V
 
 | |
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | Descriptor: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.61 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
5ZNM
 
 | | Colicin D Central Domain and C-terminal tRNase domain | | Descriptor: | Colicin-D, GLYCEROL, SULFATE ION | | Authors: | Chang, J.W, Sato, Y, Ogawa, T, Arakawa, T, Fukai, S, Fushinobu, S, Masaki, H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the central and the C-terminal RNase domains of colicin D implicated its translocation pathway through inner membrane of target cell
J. Biochem., 164, 2018
|
|
6DPV
 
 | | Undecorated GDP microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Separating the effects of nucleotide and EB binding on microtubule structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
