4RNF
 
 | |
1KX2
 
 | | Minimized average structure of a mono-heme ferrocytochrome c from Shewanella putrefaciens | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-30 | | Release date: | 2002-02-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
1J8I
 
 | | Solution Structure of Human Lymphotactin | | Descriptor: | Lymphotactin | | Authors: | Kuloglu, E.S, McCaslin, D.R, Markley, J.L, Pauza, C.D, Volkman, B.F. | | Deposit date: | 2001-05-21 | | Release date: | 2001-10-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
3BJO
 
 | |
4JMY
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with DDIVPC peptide | | Descriptor: | HCV non-structural protein 4A, Polyprotein, SODIUM ION, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Importance of the peptide scaffold of drugs that target the hepatitis C virus NS3 protease and its crucial bioactive conformation and dynamic factors.
To be Published
|
|
3RKH
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite (full occupancy) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6I4A
 
 | | Structure of P. aeruginosa LpxC with compound 18d: (2R)-N-Hydroxy-4-(6-((1-(hydroxymethyl)cyclopropyl)buta-1,3-diyn-1-yl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-[4-[1-(hydroxymethyl)cyclopropyl]buta-1,3-diynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
4OBX
 
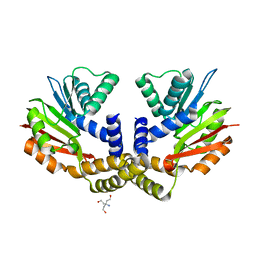 | | Crystal structure of yeast Coq5 in the apo form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5E6O
 
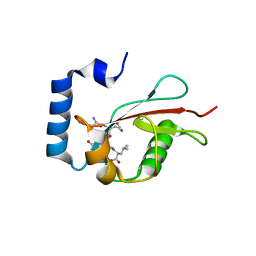 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | Descriptor: | Protein lgg-2, TRP-GLU-GLU-LEU | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
6I48
 
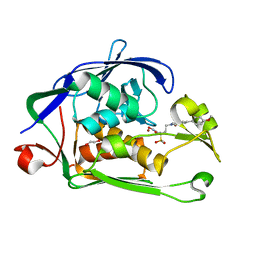 | | Structure of P. aeruginosa LpxC with compound 12: (2R)-4-(6-(2-Fluoro-4-methoxyphenyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
3QIW
 
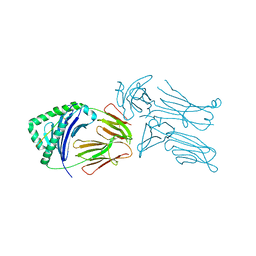 | | Crystal structure of the 226 TCR in complex with MCC-p5E/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, E-K alpha chain, ... | | Authors: | Kruse, A.C, Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
2PVR
 
 | | Crystal structure of the catalytic subunit of protein kinase CK2 (C-terminal deletion mutant 1-335) in complex with two sulfate ions | | Descriptor: | Casein kinase II subunit alpha, catalytic subunit, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Niefind, K, Yde, C.W, Ermakova, I, Issinger, O.-G. | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Evolved to Be Active: Sulfate Ions Define Substrate Recognition Sites of CK2alpha and Emphasise its Exceptional Role within the CMGC Family of Eukaryotic Protein Kinases
J.Mol.Biol., 370, 2007
|
|
5CCP
 
 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
5EDE
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c13c(cc(s1)C(NCC2OCCC2)=O)c(nn3c4ccc(cc4)Cl)C, micromolar IC50=0.217 | | Descriptor: | 1-(4-chlorophenyl)-3-methyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]thieno[2,3-c]pyrazole-5-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
4OBW
 
 | | crystal structure of yeast Coq5 in the SAM bound form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, S-ADENOSYLMETHIONINE, ... | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2JWO
 
 | | A PHD finger motif in the C-terminus of RAG2 modulates recombination activity | | Descriptor: | V(D)J recombination-activating protein 2, ZINC ION | | Authors: | Ivanov, D, Hyberts, S.G, Sun, Z, Wagner, G. | | Deposit date: | 2007-10-17 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A PHD finger motif in the C terminus of RAG2 modulates recombination activity.
J.Biol.Chem., 280, 2005
|
|
1J9O
 
 | | SOLUTION STRUCTURE OF HUMAN LYMPHOTACTIN | | Descriptor: | LYMPHOTACTIN | | Authors: | Kuloglu, E.S, McCaslin, D.R, Kitabwalla, M, Pauza, C.D, Markley, J.L, Volkman, B.F. | | Deposit date: | 2001-05-28 | | Release date: | 2001-10-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
4CDA
 
 | | Spectroscopically-validated structure of ferric cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C4Q
 
 | | Cryo-EM map of the CSFV IRES in complex with the small ribosomal 40S subunit and DHX29 | | Descriptor: | INTERNAL RIBOSOMAL ENTRY SITE | | Authors: | Hashem, Y, desGeorges, A, Dhote, V, Langlois, R, Liao, H.Y, Grassucci, R.A, Pestova, T.V, Hellen, C.U.T, Frank, J. | | Deposit date: | 2013-09-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Hepatitis-C-Virus-Like Internal Ribosome Entry Sites Displace Eif3 to Gain Access to the 40S Subunit
Nature, 503, 2013
|
|
3RNY
 
 | | Crystal structure of human RSK1 C-terminal kinase domain | | Descriptor: | Ribosomal protein S6 kinase alpha-1, SODIUM ION | | Authors: | Li, D, Fu, T.-M, Nan, J, Su, X.-D. | | Deposit date: | 2011-04-24 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the autoinhibition of the C-terminal kinase domain of human RSK1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4CCP
 
 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
1LPH
 
 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
3NNG
 
 | | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR258E | | Descriptor: | CALCIUM ION, uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Dimaio, F, Baker, D, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Crystal structure of the F5/8 type C domain of Q5LFR2_BACFN protein from Bacteroides fragilis.
To be Published
|
|
3KYG
 
 | | Crystal structure of VCA0042 (L135R) complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein VCA0042 | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
1MG7
 
 | | Crystal Structure of xol-1 | | Descriptor: | early switch protein xol-1 2.2k splice form | | Authors: | Luz, J.G, Hassig, C.A, Godzik, A, Meyer, B.J, Wilson, I.A. | | Deposit date: | 2002-08-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators
Genes Dev., 17, 2003
|
|
