6PRJ
 
 | |
6PMG
 
 | |
5J17
 
 | |
6PQT
 
 | |
5JHJ
 
 | | M. Oryzae effector AVR-Pia mutant H3 | | Descriptor: | Antivirulence protein AVR-Pia | | Authors: | Padilla, A, deGuillen, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Recognition of the Magnaporthe oryzae Effector AVR-Pia by the Decoy Domain of the Rice NLR Immune Receptor RGA5.
Plant Cell, 29, 2017
|
|
6PRI
 
 | |
6PQE
 
 | |
5JOM
 
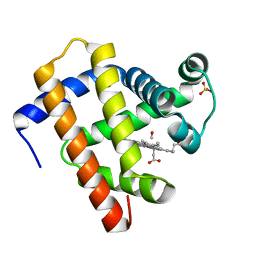 | | X-ray structure of CO-bound sperm whale myoglobin using a fixed target crystallography chip | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Oghbaey, S, Sarracini, A, Ginn, H.M, Pare-Labrosse, O, Kuo, A, Marx, A, Epp, S.W, Sherrell, D.A, Eger, B.T, Zhong, Y, Loch, R, Mariani, V, Alonso-Mori, R, Nelson, S, Lemke, H.T, Owen, R.L, Pearson, A.R, Stuart, D.I, Ernst, O.P, Mueller-Werkmeister, H.M, Miller, R.J.D. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
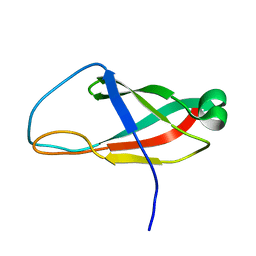
6PRP
 
 | |
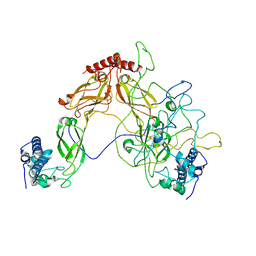
6PSI
 
 | |
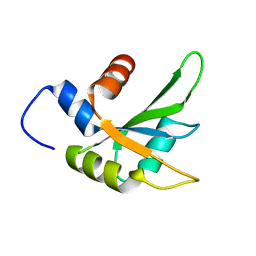
6Q2I
 
 | |
5JRG
 
 | | Crystal structure of the nucleosome containing the DNA with tetrahydrofuran (THF) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Arimura, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphism of apyrimidinic DNA structures in the nucleosome
Sci Rep, 7, 2017
|
|
6N2P
 
 | | Helical assembly of the CARD9 CARD | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Holliday, M.J, Rohou, A, Arthur, C.P, Dueber, E.C, Fairbrother, W.J. | | Deposit date: | 2018-11-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of autoinhibited and polymerized forms of CARD9 reveal mechanisms of CARD9 and CARD11 activation.
Nat Commun, 10, 2019
|
|
5JTL
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
7JK3
 
 | | Structure of Drosophila ORC bound to GC-rich DNA and Cdc6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein, DNA (33-MER), ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
5JLH
 
 | | Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 2, ... | | Authors: | von der Ecken, J, Heissler, S.M, Pathan-Chhatbar, S, Manstein, D.J, Raunser, S. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution.
Nature, 534, 2016
|
|
5IYU
 
 | | AlgE_CIM | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alginate production protein AlgE, ... | | Authors: | Ma, P, Weichert, D. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
5XOB
 
 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
5JTN
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site c | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
6S3S
 
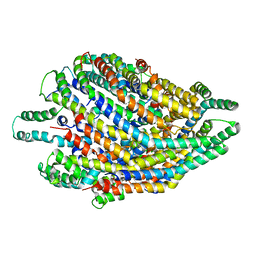 | | Structure of the FliPQR complex from the flagellar type 3 secretion system of Vibrio mimicus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Kuhlen, L, Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion.
Nat Commun, 11, 2020
|
|
5JYG
 
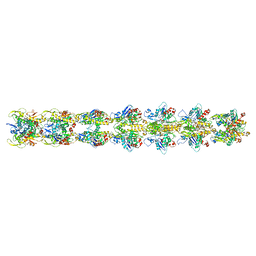 | |
5HI6
 
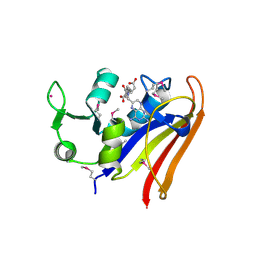 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
4QXK
 
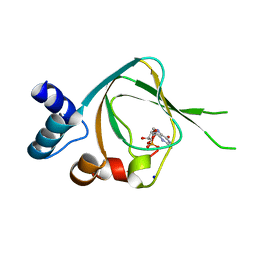 | | Joint X-ray/neutron structure of PKGIbeta in complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, SODIUM ION, cGMP-dependent protein kinase 1 | | Authors: | Kim, C, Gerlits, O, Kovalevsky, A, Huang, G.Y. | | Deposit date: | 2014-07-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Neutron Diffraction Reveals Hydrogen Bonds Critical for cGMP-Selective Activation: Insights for cGMP-Dependent Protein Kinase Agonist Design.
Biochemistry, 53, 2014
|
|
5JZH
 
 | | Cryo-EM structure of aerolysin prepore | | Descriptor: | Aerolysin | | Authors: | Iacovache, I, Zuber, B. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process.
Nat Commun, 7, 2016
|
|
5K5E
 
 | | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | De la Mora, E, Dighe, S.N, Deora, G.S, Ross, B.P, Nachon, F, Brazzolotto, X. | | Deposit date: | 2016-05-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening.
J.Med.Chem., 59, 2016
|
|
