5JZH
 
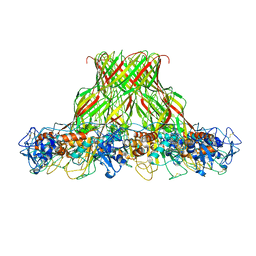 | | Cryo-EM structure of aerolysin prepore | | Descriptor: | Aerolysin | | Authors: | Iacovache, I, Zuber, B. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-13 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process.
Nat Commun, 7, 2016
|
|
7Q9Y
 
 | |
5JZW
 
 | |
5JZT
 
 | |
