4GV9
 
 | | Lassa nucleoprotein C-terminal domain in complex with triphosphated dsRNA soaking for 5 min | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, RNA (5'-R(*(GTP)P*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
2PVR
 
 | | Crystal structure of the catalytic subunit of protein kinase CK2 (C-terminal deletion mutant 1-335) in complex with two sulfate ions | | Descriptor: | Casein kinase II subunit alpha, catalytic subunit, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Niefind, K, Yde, C.W, Ermakova, I, Issinger, O.-G. | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Evolved to Be Active: Sulfate Ions Define Substrate Recognition Sites of CK2alpha and Emphasise its Exceptional Role within the CMGC Family of Eukaryotic Protein Kinases
J.Mol.Biol., 370, 2007
|
|
2MTS
 
 | | Three-Dimensional Structure and Interaction Studies of Hepatitis C Virus p7 in 1,2-Dihexanoyl-sn-glycero-3-phosphocholine by Solution Nuclear Magnetic Resonance | | Descriptor: | HEPATITIS C VIRUS P7 PROTEIN | | Authors: | Cook, G.A, Dawson, L.A, Tian, Y, Opella, S.J. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance.
Biochemistry, 52, 2013
|
|
4HXK
 
 | | Brd4 Bromodomain 1 complex with 6,7-DIHYDROTHIENO[3,2-C]PYRIDIN-5(4H)-YL(1H-IMIDAZOL-1-YL)METHANONE inhibitor | | Descriptor: | 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl(1H-imidazol-1-yl)methanone, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
2K90
 
 | | Dimeric solution structure of the DNA loop d(TGCTTCGT) | | Descriptor: | 5'-D(*TP*GP*CP*TP*TP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2K97
 
 | | Dimeric solution structure of the cyclic octamer d(pCGCTCCGT) | | Descriptor: | 5'-D(P*CP*GP*CP*TP*CP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-30 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
1CXC
 
 | | CRYSTALLIZATION AND X-RAY STRUCTURE DETERMINATION OF CYTOCHROME C2 FROM RHODOBACTER SPHAEROIDES IN THREE CRYSTAL FORMS | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Axelrod, H.L, Feher, G, Allen, J.P, Chirino, A.J, Day, M.W, Hsu, B.T, Rees, D.C. | | Deposit date: | 1994-02-14 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization and X-ray structure determination of cytochrome c2 from Rhodobacter sphaeroides in three crystal forms.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
3WWT
 
 | | Crystal Structure of the Y3:STAT1ND complex | | Descriptor: | C' protein, CALCIUM ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Oda, K, Sakaguchi, T, Matoba, Y. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of the Inhibition of STAT1 Activity by Sendai Virus C Protein.
J.Virol., 89, 2015
|
|
2A3M
 
 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome (oxidized form) | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C | | Authors: | Pattarkine, M.V, Tanner, J.J, Bottoms, C.A, Lee, Y.H, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
6IQR
 
 | |
7CCP
 
 | |
2L19
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Arsenate reductase | | Authors: | Yu, C, Xia, B, Jin, C. | | Deposit date: | 2010-07-26 | | Release date: | 2011-04-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C and (15)N resonance assignments of the arsenate reductase from Synechocystis sp. strain PCC 6803
Biomol.Nmr Assign., 5, 2011
|
|
8GBT
 
 | | Time-resolve SFX structure of a photoproduct of carbon monoxide complex of bovine cytochrome c oxidase | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Yeh, S.-R, Rousseau, D.L. | | Deposit date: | 2023-02-28 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detection of a Geminate Photoproduct of Bovine Cytochrome c Oxidase by Time-Resolved Serial Femtosecond Crystallography.
J.Am.Chem.Soc., 145, 2023
|
|
2XTS
 
 | | Crystal Structure of the Sulfane Dehydrogenase SoxCD from Paracoccus pantotrophus | | Descriptor: | CALCIUM ION, COBALT (II) ION, CYTOCHROME, ... | | Authors: | Zander, U, Faust, A, Klink, B.U, de Sanctis, D, Panjikar, S, Quentmeier, A, Bardischewski, F, Friedrich, C.G, Scheidig, A.J. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural Basis for the Oxidation of Protein-Bound Sulfur by the Sulfur Cycle Molybdohemo-Enzyme Sulfane Dehydrogenase Soxcd.
J.Biol.Chem., 286, 2011
|
|
3SW0
 
 | | Structure of the C-terminal region (modules 18-20) of complement regulator Factor H | | Descriptor: | Complement factor H, GLYCEROL, PHOSPHATE ION | | Authors: | Morgan, H.P, Guariento, M, Schmidt, C.Q, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of the C-Terminal Region (Modules 18-20) of Complement Regulator Factor H (FH).
Plos One, 7, 2012
|
|
2VUV
 
 | | Crystal structure of Codakine at 1.3A resolution | | Descriptor: | CALCIUM ION, CITRIC ACID, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
1H9O
 
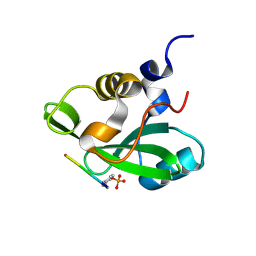 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, CRYSTAL STRUCTURE AT 1.79 A | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Pauptit, R.A, Rowsell, S, Breeze, A.L, Murshudov, G.N, Dennis, C.A, Derbyshire, D.J, Weston, S.A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2BC0
 
 | |
2BCP
 
 | |
2BC1
 
 | |
2QDQ
 
 | | Crystal structure of the talin dimerisation domain | | Descriptor: | Talin-1 | | Authors: | Gingras, A.R, Putz, N.S.M, Bate, N, Barsukov, I.L, Critchley, D.R.C. | | Deposit date: | 2007-06-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2008
|
|
1N96
 
 | | DIMERIC SOLUTION STRUCTURE OF THE CYCLIC OCTAMER CD(CGCTCATT) | | Descriptor: | CYCLIC OLIGONUCLEOTIDE D(CGCTCATT) | | Authors: | Escaja, N, Gelpi, J.L, Orozco, M, Rico, M, Pedroso, E, Gonzalez, C. | | Deposit date: | 2002-11-22 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Four-stranded DNA structure stabilized by a novel G:C:A:T tetrad.
J.Am.Chem.Soc., 125, 2003
|
|
1N9C
 
 | |
3G6H
 
 | | Src Thr338Ile inhibited in the DFG-Asp-Out conformation | | Descriptor: | N-{4-methyl-3-[(3-{4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
2YQB
 
 | | Structure of P93A variant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.4 A resolution | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Han, C, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
