4OVI
 
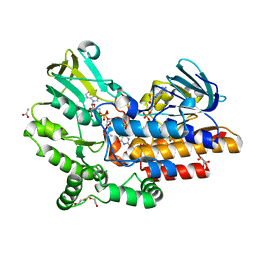 | | Phenylacetone monooxygenase: oxidised enzyme in complex with APADP | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martinoli, C, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Beyond the Protein Matrix: Probing Cofactor Variants in a Baeyer-Villiger Oxygenation Reaction.
Acs Catalysis, 3, 2013
|
|
4OSO
 
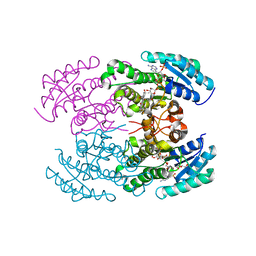 | | The crystal structure of landomycin C-6 ketoreductase LanV with bound NADP and rabelomycin | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reductase homolog, ... | | Authors: | Paananen, P, Niiranen, L, Patrikainen, P, Metsa-Ketela, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based engineering of angucyclinone 6-ketoreductases.
Chem.Biol., 21, 2014
|
|
4EHB
 
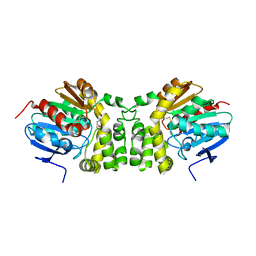 | |
4EUS
 
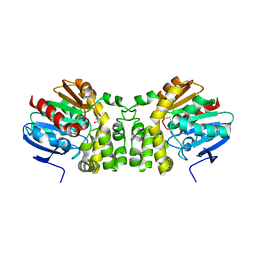 | |
7OWB
 
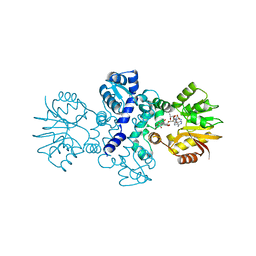 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-hydroxylase CalMB | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
6LUT
 
 | |
3TTQ
 
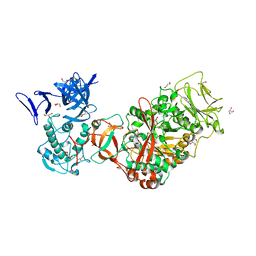 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
4EX7
 
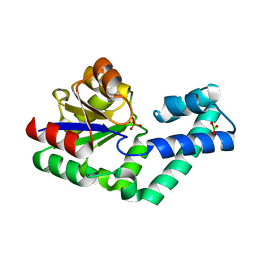 | | Crystal structure of the alnumycin P phosphatase in complex with free phosphate | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX8
 
 | | Crystal structure of the prealnumycin C-glycosynthase AlnA | | Descriptor: | AlnA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UIB
 
 | | Map kinase LMAMPK10 from leishmania major in complex with SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, mitogen-activated protein kinase | | Authors: | Horjales, S, Schmidt-Arras, D, Leclercq, O, Spath, G, Buschiazzo, A. | | Deposit date: | 2011-11-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of the MAP Kinase LmaMPK10 from Leishmania Major Reveals Parasite-Specific Features and Regulatory Mechanisms.
Structure, 20, 2012
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
4EX9
 
 | | Crystal structure of the prealnumycin C-glycosynthase AlnA in complex with ribulose 5-phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AlnA, CALCIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7PGJ
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-decarboxylate TamK and 10-hydroxylase RdmB, together with a single point mutation F297G | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-7-methoxy-2,4,5-tris(oxidanyl)-6,11-bis(oxidanylidene)-3,4-dihydro-1H-tetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PG7
 
 | |
7PHF
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PGA
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PHD
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | Authors: | Grocholski, T, Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PHE
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
1I1I
 
 | | NEUROLYSIN (ENDOPEPTIDASE 24.16) CRYSTAL STRUCTURE | | Descriptor: | NEUROLYSIN, ZINC ION | | Authors: | Brown, C.K, Madauss, K, Lian, W, Tolbert, W.D, Beck, M.R, Rodgers, D.W. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of neurolysin reveals a deep channel that limits substrate access.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3TFH
 
 | | DMSP-dependent demethylase from P. ubique - apo | | Descriptor: | GLYCEROL, GcvT-like Aminomethyltransferase protein, SODIUM ION | | Authors: | Schuller, D.J, Reisch, C.R, Moran, M.A, Whitman, W.B, Lanzilotta, W.N. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagabacter ubique.
Protein Sci., 21, 2012
|
|
3TFJ
 
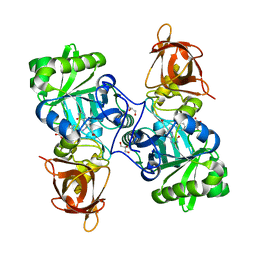 | | DMSP-dependent demethylase from P. ubique - with cofactor THF | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, ACETATE ION, GLYCEROL, ... | | Authors: | Schuller, D.J, Reisch, C.R, Moran, M.A, Whitman, W.B, Lanzilotta, W.N. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagabacter ubique.
Protein Sci., 21, 2012
|
|
3TTO
 
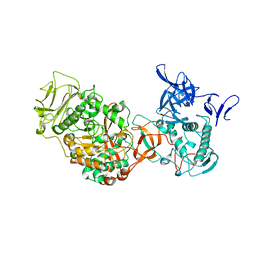 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in triclinic form | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Tranier, S, Remaud-Simeon, M, Dijkstra, B.W. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
4HDD
 
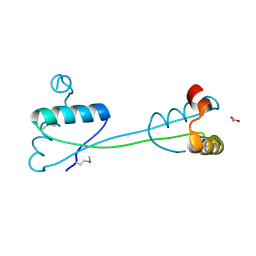 | | Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease | | Descriptor: | ACETATE ION, Rhomboid protease GlpG | | Authors: | Lazareno-Saez, C, Arutyunova, E, Coquelle, N, Lemieux, M.J. | | Deposit date: | 2012-10-02 | | Release date: | 2013-02-06 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease.
J.Mol.Biol., 425, 2013
|
|
3ZVH
 
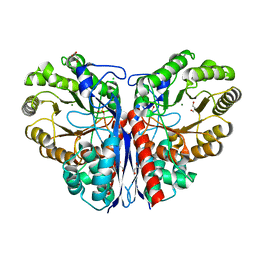 | | Methylaspartate ammonia lyase from Clostridium tetanomorphum mutant Q73A | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Raj, H, Szymanski, W, de Villiers, J, Rozeboom, H.J, Veetil, V.P, Reis, C.R, de Villiers, M, de Wildeman, S, Dekker, F.J, Quax, W.J, Thunnissen, A.M.W.H, Feringa, B.L, Janssen, D.B, Poelarends, G.J. | | Deposit date: | 2011-07-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Nat.Chem., 4, 2012
|
|
4R9K
 
 | | Structure of thermostable eightfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis | | Descriptor: | (2R)-2-hydroxyhexanamide, GLYCEROL, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
