3SQV
 
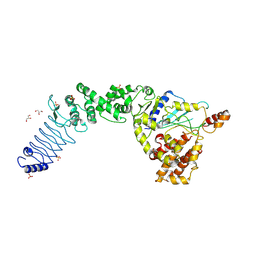 | | Crystal Structure of E. coli O157:H7 E3 ubiquitin ligase, NleL, with a human E2, UbcH7 | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Lin, D.Y, Chen, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of two bacterial HECT-like E3 ligases in complex with a human E2 reveal atomic details of pathogen-host interactions.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5UBW
 
 | |
8ST9
 
 | |
8JZO
 
 | |
8JCA
 
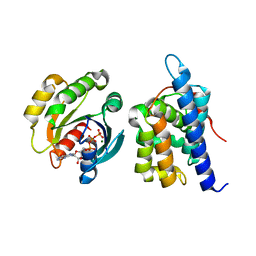 | |
2ML9
 
 | |
9F14
 
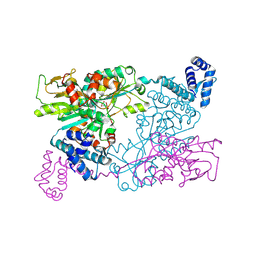 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
9FDD
 
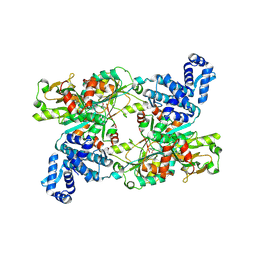 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-05-16 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
1K3E
 
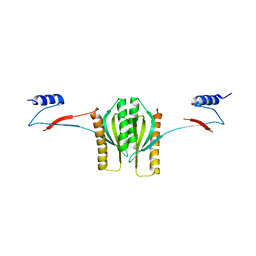 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
2XGA
 
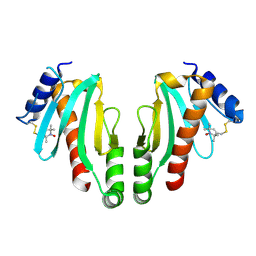 | | MTSL spin-labelled Shigella Flexneri Spa15 | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, SURFACE PRESENTATION OF ANTIGENS PROTEIN SPAK | | Authors: | Lillington, J.E.D, Johnson, S, Lea, S.M. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shigella Flexneri Spa15 Crystal Structure Verified in Solution by Double Electron Electron Resonance.
J.Mol.Biol., 405, 2011
|
|
2VT1
 
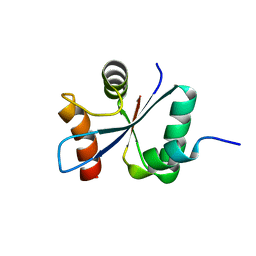 | | Crystal structure of the cytoplasmic domain of Spa40, the specificity switch for the Shigella flexneri Type III Secretion System | | Descriptor: | SURFACE PRESENTATION OF ANTIGENS PROTEIN SPAS | | Authors: | Deane, J.E, Graham, S.C, Mitchell, E.P, Flot, D, Johnson, S, Lea, S.M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Spa40, the Specificity Switch for the Shigella Flexneri Type III Secretion System
Mol.Microbiol., 69, 2008
|
|
2VRW
 
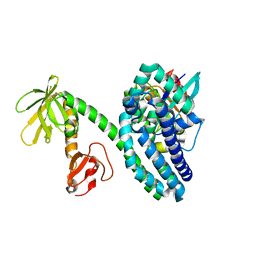 | |
3GCG
 
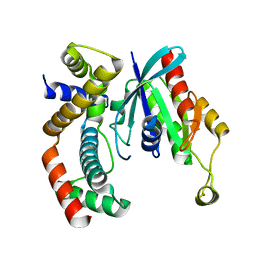 | | crystal structure of MAP and CDC42 complex | | Descriptor: | Cell division control protein 42 homolog, L0028 (Mitochondria associated protein) | | Authors: | Chai, J, Huang, Z, Feng, Y, Wu, X. | | Deposit date: | 2009-02-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics
Nat.Struct.Mol.Biol., 16, 2009
|
|
1YJ7
 
 | | Crystal structure of enteropathogenic E.coli (EPEC) type III secretion system protein EscJ | | Descriptor: | GLYCEROL, PHOSPHATE ION, escJ | | Authors: | Yip, C.K, Kimbrough, T.G, Felise, H.B, Vuckovic, M, Thomas, N.A, Pfuetzner, R.A, Frey, E.A, Finlay, B.B, Miller, S.I, Strynadka, N.C.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the molecular platform for type III secretion system assembly.
Nature, 435, 2005
|
|
2WKR
 
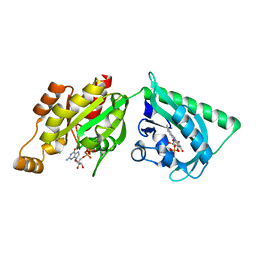 | | Structure of a photoactivatable Rac1 containing the Lov2 C450M Mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
2WKP
 
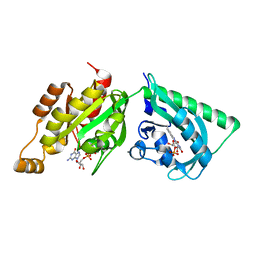 | | Structure of a photoactivatable Rac1 containing Lov2 Wildtype | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
2WKQ
 
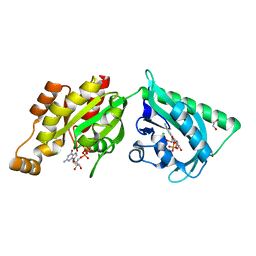 | | Structure of a photoactivatable Rac1 containing the Lov2 C450A Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, Y.I, Frey, D, Lungu, O.I, Jaehrig, A, Schlichting, I, Kuhlman, B, Hahn, K.M. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells.
Nature, 461, 2009
|
|
2YIN
 
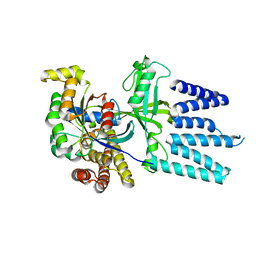 | | STRUCTURE OF THE COMPLEX BETWEEN Dock2 AND Rac1. | | Descriptor: | DEDICATOR OF CYTOKINESIS PROTEIN 2, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 1 | | Authors: | Kulkarni, K.A, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple Factors Confer Specific Cdc42 and Rac Protein Activation by Dedicator of Cytokinesis (Dock) Nucleotide Exchange Factors.
J.Biol.Chem., 286, 2011
|
|
2MXQ
 
 | | The solution structure of DEFA1, a highly potent antimicrobial peptide from the horse | | Descriptor: | Paneth cell-specific alpha-defensin 1 | | Authors: | Jung, S, Michalek, M, Shomali, M, Soennichsen, F.D. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional studies of the highly potent equine antimicrobial peptide DEFA1.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
3U0C
 
 | |
