7PZA
 
 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
8IDW
 
 | |
3IED
 
 | | Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF14_0417) in complex with AMPPN | | Descriptor: | AMP PHOSPHORAMIDATE, Heat shock protein | | Authors: | Pizarro, J.C, Wernimont, A.K, Lew, J, Hutchinson, A, Artz, J.D, Amaya, M.F, Plotnikova, O, Vedadi, M, Kozieradzki, I, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Botchkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF14_0417) in complex with AMPPN
TO BE PUBLISHED
|
|
8VUX
 
 | | Cryo-EM structure of human ABC transporter (hABCC1) bound to cGAMP | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Multidrug resistance-associated protein 1, cGAMP | | Authors: | Shinde, O, Li, P. | | Deposit date: | 2024-01-30 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of ATP-binding cassette transporter ABCC1 reveal the molecular basis of cyclic dinucleotide cGAMP export.
Immunity, 58, 2025
|
|
7MYJ
 
 | | Structure of full length human AMPK (a2b1g1) in complex with a small molecule activator MSG011 | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Ovens, A.J, Gee, Y.S, Ling, N.X.Y, Waters, N.J, Yu, D, Scott, J.W, Parker, M.W, Hoffman, N.J, Kemp, B.E, Baell, J.B, Oakhill, J.S, Langendorf, C.G. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-function analysis of the AMPK activator SC4 and identification of a potent pan AMPK activator.
Biochem.J., 479, 2022
|
|
1T8W
 
 | |
1T8S
 
 | |
1T8Y
 
 | |
1T8R
 
 | |
6EOF
 
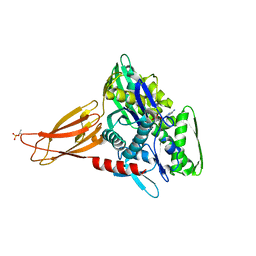 | | Crystal structure of AMPylated GRP78 in ADP state | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, Y, Preissler, S, Read, R.J, Ron, D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
8SI0
 
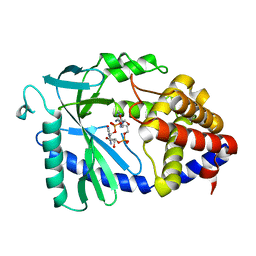 | |
5O4P
 
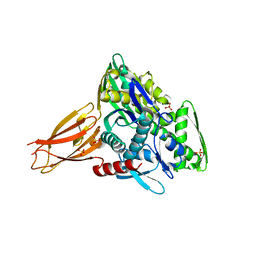 | | Crystal structure of AMPylated GRP78 | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Yan, Y, Chen, R, Ron, D, Read, R. | | Deposit date: | 2017-05-30 | | Release date: | 2017-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
8T00
 
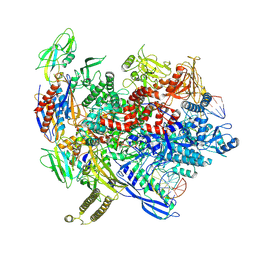 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: closed duplex DNA (rPTCc) | | Descriptor: | DNA (26-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-31 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8T0L
 
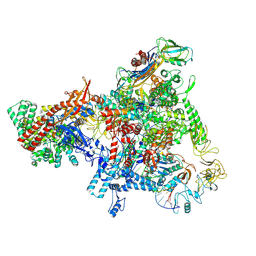 | | E. coli Sw2/Snf2 ATPase RapA bound to both ADP-AlF3 and reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA (29-MER), ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-06-01 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7MWZ
 
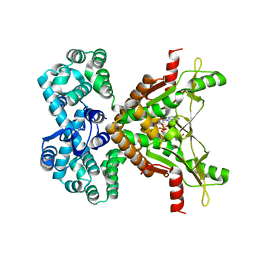 | |
1STC
 
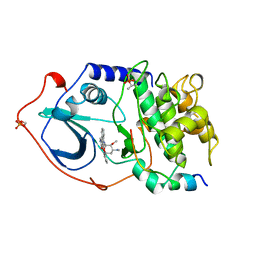 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
1USQ
 
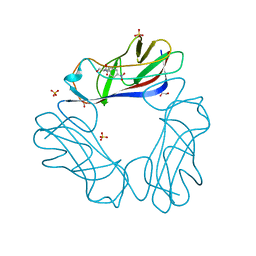 | | Complex of E. Coli DraE adhesin with Chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, ... | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-11-27 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
8T02
 
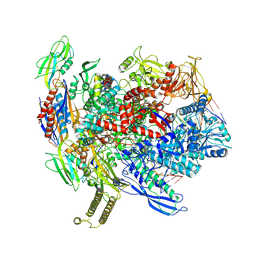 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: unwinding duplex DNA (rPTCi) | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-31 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
2XAT
 
 | |
3NU3
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NU6
 
 | |
3NU5
 
 | | Crystal Structure of HIV-1 Protease Mutant I50V with Antiviral Drug Amprenavir | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wang, Y.-F, Shen, C.H, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3O2A
 
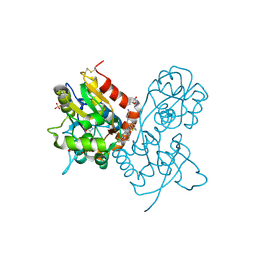 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O6I
 
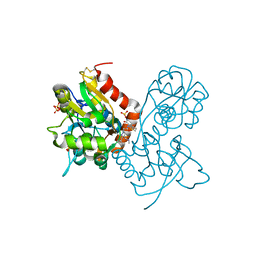 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-[({3-tert-butyl-4-[(methylamino)methyl]-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2JKL
 
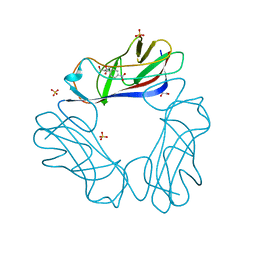 | | DraE Adhesin in complex with Bromamphenicol | | Descriptor: | 1,2-ETHANEDIOL, BROMAMPHENICOL, CHLORAMPHENICOL, ... | | Authors: | Pettigrew, D.M, Roversi, P, Davies, S.G, Russell, A.J, Lea, S.M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Study of the Interaction between the Dr Haemagglutinin Drae and Derivatives of Chloramphenicol
Acta Crystallogr.,Sect.D, 65, 2009
|
|
