6V08
 
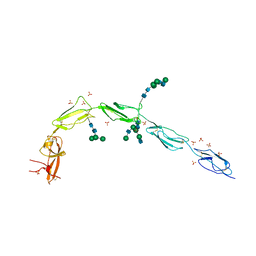 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6FCO
 
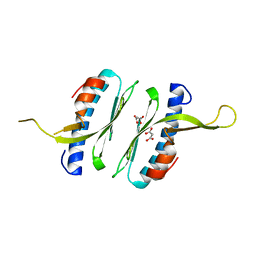 | | Structural and functional characterisation of Frataxin (FXN) like protein from Chaetomium thermophilum | | Descriptor: | MALONIC ACID, Mitochondrial frataxin-like protein | | Authors: | Jamshidiha, M, Rasheed, M, Pastore, A, Cota, E. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and functional characterization of a frataxin from a thermophilic organism.
FEBS J., 286, 2019
|
|
6V1B
 
 | | Crystal structure of the bromodomain of human BRD9 bound to I-BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
2G4B
 
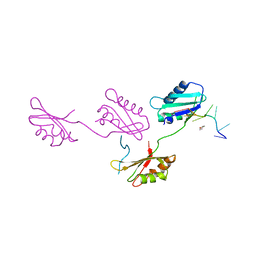 | | Structure of U2AF65 variant with polyuridine tract | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', Splicing factor U2AF 65 kDa subunit | | Authors: | Sickmier, E.A, Kielkopf, C.L. | | Deposit date: | 2006-02-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of polypyrimidine tract recognition
by the essential splicing factor U2AF65.
Mol.Cell, 23, 2006
|
|
4I9Y
 
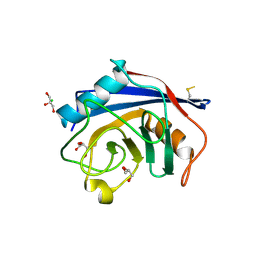 | | Structure of the C-terminal domain of Nup358 | | Descriptor: | CHLORIDE ION, E3 SUMO-protein ligase RanBP2, GLYCEROL, ... | | Authors: | Lin, D.H, Zimmermann, S, Stuwe, T, Stuwe, E, Hoelz, A. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Analysis of the C-Terminal Domain of Nup358/RanBP2.
J.Mol.Biol., 425, 2013
|
|
1DOG
 
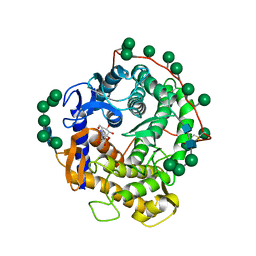 | | REFINED STRUCTURE FOR THE COMPLEX OF 1-DEOXYNOJIRIMYCIN WITH GLUCOAMYLASE FROM (ASPERGILLUS AWAMORI) VAR. X100 TO 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 1-DEOXYNOJIRIMYCIN, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Harris, E, Aleshin, A, Firsov, L, Honzatko, R.B. | | Deposit date: | 1993-01-12 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of 1-deoxynojirimycin with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
Biochemistry, 32, 1993
|
|
8DDJ
 
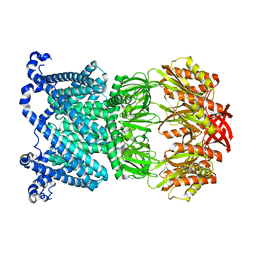 | | Open MscS in PC14.1 Nanodiscs | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Reddy, B.G, Bavi, N, Perozo, E. | | Deposit date: | 2022-06-18 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | State-specific morphological deformations of the lipid bilayer explain mechanosensitive gating of MscS ion channels.
Elife, 12, 2023
|
|
6V9K
 
 | |
2G1S
 
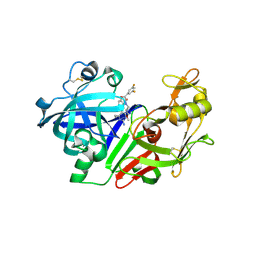 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the C Ring | | Descriptor: | (2S)-6-(2,4-DIAMINO-6-ETHYLPYRIMIDIN-5-YL)-2-(3,5-DIFLUOROPHENYL)-4-(3-METHOXYPROPYL)-2H-1,4-BENZOXAZIN-3(4H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Holsworth, D.D, Jalaiea, M, Zhanga, E, Mcconnella, P. | | Deposit date: | 2006-02-14 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
6V8O
 
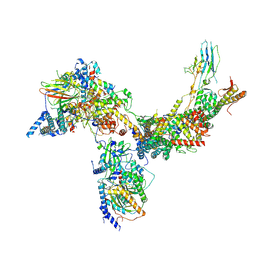 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
1HB2
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN EXPOSED PRODUCT FROM ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1S)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOCARBONYL)-2-OXOETHYL]-6-OXO-L-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
6VHA
 
 | | Singlet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
7AKP
 
 | | Crystal structure of E. coli RNA helicase HrpA-D305A | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8DGD
 
 | | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GAF domain-containing protein, ZINC ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Delker, S.L, Weiss, M.J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-06-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae
to be published
|
|
1H4D
 
 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
2GD3
 
 | | NMR structure of S14G-humanin in 30% TFE solution | | Descriptor: | Humanin | | Authors: | Benaki, D, Zikos, C, Evangelou, A, Livaniou, E, Vlassi, M, Mikros, E, Pelecanou, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ser14Gly-humanin, a potent rescue factor against neuronal cell death in Alzheimer's disease.
Biochem.Biophys.Res.Commun., 349, 2006
|
|
5C4K
 
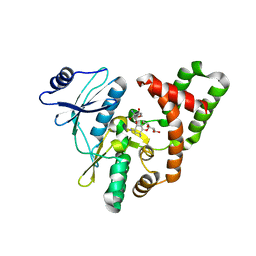 | | APH(2")-IVa in complex with GET (G418) at room temperature | | Descriptor: | APH(2'')-Id, GENETICIN | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
4DTL
 
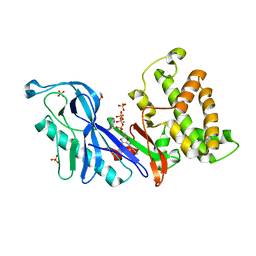 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++
J.Biol.Chem., 2012
|
|
5TLX
 
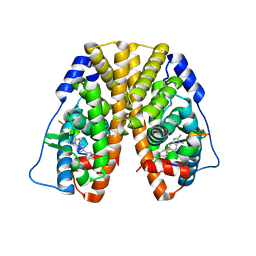 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 3,4-bis(4-hydroxyphenyl)thiophene 1,1-dioxide | | Descriptor: | 3,4-bis(4-hydroxyphenyl)-2,5-dihydro-1H-1lambda~6~-thiophene-1,1-dione, 3-methyl-6-phenyl-3H-imidazo[4,5-b]pyridin-2-amine, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6VGV
 
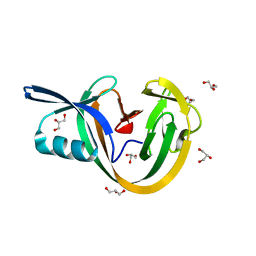 | | Crystal structure of VidaL intein | | Descriptor: | GLYCEROL, VidaL | | Authors: | Burton, A.J, Haugbro, M, Parisi, E, Muir, T.W. | | Deposit date: | 2020-01-09 | | Release date: | 2020-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Live-cell protein engineering with an ultra-short split intein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TM6
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)hexanoic acid | | Descriptor: | 6-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}hexanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4Q6E
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[3-(3,5-Dimethyl-1H-pyrazol-1-yl)-3-oxopropyl]amino}benzene-1-sulfonamide | | Descriptor: | 4-{[3-(3,5-dimethyl-1H-pyrazol-1-yl)-3-oxopropyl]amino}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 4-Amino-substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Molecules, 19, 2014
|
|
5TM5
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 5-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | Descriptor: | 5-{4-[(1S,4S,5R)-5-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-((1,3-dihydro-2H-inden-2-ylidene)methylene)diphenol | | Descriptor: | 4,4'-[(1,3-dihydro-2H-inden-2-ylidene)methylene]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN6
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Spiro BC-estradiol, (1S,1'S,3a'S,7a'S)-7a'-methyl-1',2,2',3,3',3a',4',6',7',7a'-decahydro-1,5'-spirobi[indene]-1',5-diol | | Descriptor: | (1S,1'S,3a'S,7a'S)-7a'-methyl-1',2,2',3,3',3a',4',6',7',7a'-decahydro-1,5'-spirobi[indene]-1',5-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
