5HUI
 
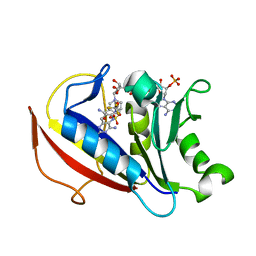 | | 6-substituted pyrido[3,2-d]pyrimidine--6-4'-trifluoromethoxyphenyl) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-[4-(trifluoromethoxy)phenyl]pyrido[3,2-d]pyrimidine-2,4,6-triamine, ... | | Authors: | Cody, V. | | Deposit date: | 2016-01-27 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Human dihydrofolate reductase ternary complex with a series of 6-substituted pyrido[3,2]pyrimidine-6-(4'-trifluoromethoxypheny)-2,4-diamines
To Be Published
|
|
2D41
 
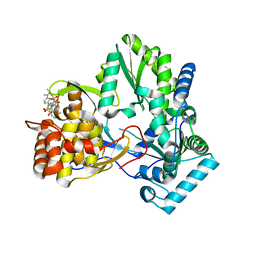 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | Descriptor: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-05 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3Z
 
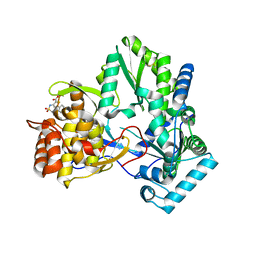 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2QA4
 
 | |
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6XAA
 
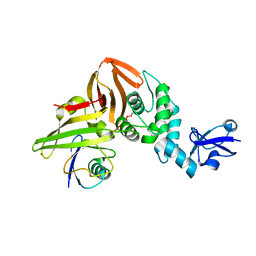 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
11BA
 
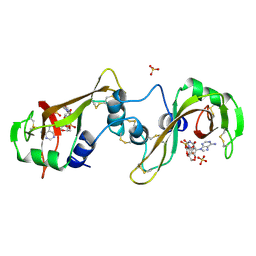 | | BINDING OF A SUBSTRATE ANALOGUE TO A DOMAIN SWAPPING PROTEIN IN THE COMPLEX OF BOVINE SEMINAL RIBONUCLEASE WITH URIDYLYL-2',5'-ADENOSINE | | Descriptor: | PROTEIN (RIBONUCLEASE, SEMINAL), SULFATE ION, ... | | Authors: | Vitagliano, L, Adinolfi, S, Riccio, A, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-17 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of a substrate analog to a domain swapping protein: X-ray structure of the complex of bovine seminal ribonuclease with uridylyl(2',5')adenosine.
Protein Sci., 7, 1998
|
|
2D3U
 
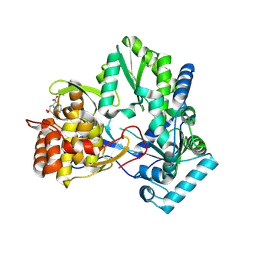 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
1MIM
 
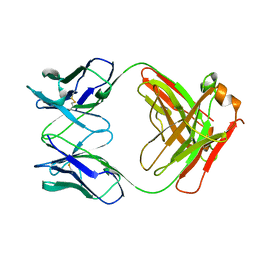 | | IGG FAB FRAGMENT (CD25-BINDING) | | Descriptor: | CHIMERIC SDZ CHI621 | | Authors: | Mikol, V. | | Deposit date: | 1995-12-04 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fab fragment of SDZ CHI621: a chimeric antibody against CD25.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5IFG
 
 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
6XF1
 
 | |
1MV8
 
 | | 1.55 A crystal structure of a ternary complex of GDP-mannose dehydrogenase from Psuedomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, GDP-mannose 6-dehydrogenase, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of GDP-mannose
dehydrogenase: A key enzyme in alginate
biosynthesis of P. aeruginosa
Biochemistry, 42, 2003
|
|
8C4H
 
 | | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly multimer | | Descriptor: | Nucleocapsid, RNA (84-MER) | | Authors: | Passchier, T.C, Maskell, D.P, Edwards, T.A, Barr, J.N. | | Deposit date: | 2023-01-04 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.485 Å) | | Cite: | The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Sci Rep, 14, 2024
|
|
8CBW
 
 | | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly monomer | | Descriptor: | Nucleocapsid, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Passchier, T.C, Maskell, D.P, Edwards, T.A, Barr, J.N. | | Deposit date: | 2023-01-26 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.485 Å) | | Cite: | The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Sci Rep, 14, 2024
|
|
1OKS
 
 | | Crystal structure of the measles virus phosphoprotein XD domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Johansson, K, Bourhis, J.-M, Campanacci, V, Cambillau, C, Canard, B, Longhi, S. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Measles Virus Phosphoprotein Domain Responsible for the Induced Folding of the C-Terminal Domain of the Nucleoprotein
J.Biol.Chem., 278, 2003
|
|
6XF2
 
 | |
6XKQ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
1WBU
 
 | | Fragment based lead discovery using crystallography | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, RIBONUCLEASE | | Authors: | Cleasby, A, Hartshorn, M.J, Murray, C.W, Jhoti, H, Tickle, I.J. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
6XO4
 
 | |
6XKP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6V53
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000985494 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000985494
To Be Published
|
|
1P5N
 
 | |
5MG2
 
 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
8D9D
 
 | | Human DNA polymerase-alpha/primase elongation complex II bound to primer/template | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*TP*GP*GP*TP*CP*GP*TP*GP*CP*CP*GP*CP*CP*AP*AP*TP*AP*A)-3'), DNA polymerase alpha catalytic subunit, ... | | Authors: | He, Q, Baranovskiy, A, Lim, C, Tahirov, T. | | Deposit date: | 2022-06-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structures of human primosome elongation complexes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3KK1
 
 | |
