2JB5
 
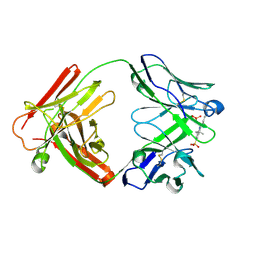 | | Fab fragment in complex with small molecule hapten, crystal form-1 | | Descriptor: | 2-{(1E,3Z,5E,7E)-7-[3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-1,3-DIHYDRO-2H-INDOL-2-YLIDENE]-4-METHYLHEPTA-1,3,5-TRIEN-1-YL}-3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-3H-INDOLIUM, FAB FRAGMENT MOR03268 HEAVY CHAIN, FAB FRAGMENT MOR03268 LIGHT CHAIN | | Authors: | Hillig, R.C, Baesler, S, Malawski, G, Badock, V, Bahr, I, Schirner, M, Licha, K. | | Deposit date: | 2006-12-03 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fab Mor03268 Triggers Absorption Shift of a Diagnostic Dye Via Packaging in a Solvent-Shielded Fab Dimer Interface
J.Mol.Biol., 377, 2008
|
|
1HYV
 
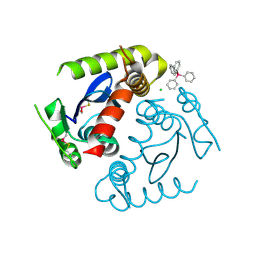 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH TETRAPHENYL ARSONIUM | | Descriptor: | CHLORIDE ION, INTEGRASE, SULFATE ION, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HYZ
 
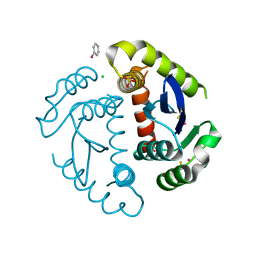 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH A DERIVATIVE OF TETRAPHENYL ARSONIUM. | | Descriptor: | (3,4-DIHYDROXY-PHENYL)-TRIPHENYL-ARSONIUM, CHLORIDE ION, INTEGRASE, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3GWL
 
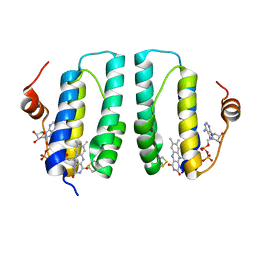 | |
3GWN
 
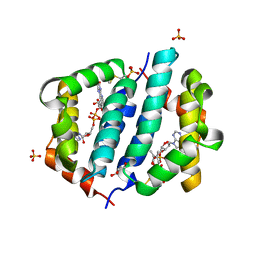 | |
1LKY
 
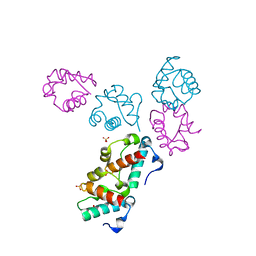 | | Structure of the wild-type TEL-SAM polymer | | Descriptor: | SULFATE ION, TRANSCRIPTION FACTOR ETV6 | | Authors: | Tran, H.H, Kim, C.A, Faham, S, Bowie, J.U. | | Deposit date: | 2002-04-26 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Native interface of the SAM domain polymer of TEL.
BMC STRUCT.BIOL., 2, 2002
|
|
1B6S
 
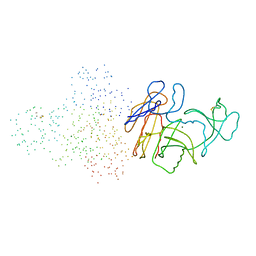 | | STRUCTURE OF N5-CARBOXYAMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (N5-CARBOXYAMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE) | | Authors: | Thoden, J.B, Kappock, T.J, Stubbe, J, Holden, H.M. | | Deposit date: | 1999-01-18 | | Release date: | 1999-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of N5-carboxyaminoimidazole ribonucleotide synthetase: a member of the ATP grasp protein superfamily.
Biochemistry, 38, 1999
|
|
5L0O
 
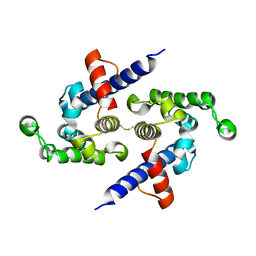 | |
1B6R
 
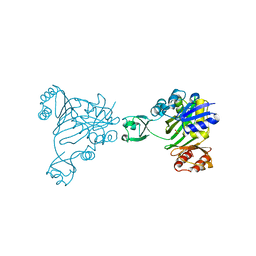 | | N5-CARBOXYAMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE FROM E. COLI | | Descriptor: | PROTEIN (N5-CARBOXYAMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE), SULFATE ION | | Authors: | Thoden, J.B, Kappock, T.J, Stubbe, J, Holden, H.M. | | Deposit date: | 1999-01-17 | | Release date: | 1999-11-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of N5-carboxyaminoimidazole ribonucleotide synthetase: a member of the ATP grasp protein superfamily.
Biochemistry, 38, 1999
|
|
4OM7
 
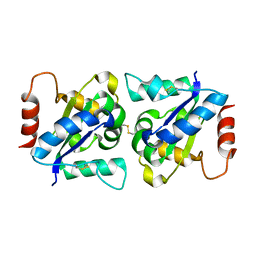 | | Crystal structure of TIR domain of TLR6 | | Descriptor: | Toll-like receptor 6 | | Authors: | Park, H.H, Jang, T.H. | | Deposit date: | 2014-01-26 | | Release date: | 2014-08-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal Structure of TIR Domain of TLR6 Reveals Novel Dimeric Interface of TIR-TIR Interaction for Toll-Like Receptor Signaling Pathway.
J.Mol.Biol., 426, 2014
|
|
7AED
 
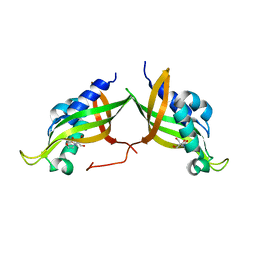 | | VirB8 domain of PrgL from Enterococcus faecalis pCF10 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PrgL | | Authors: | Jaeger, F, Berntsson, R.P.A. | | Deposit date: | 2020-09-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structure of the enterococcal T4SS protein PrgL reveals unique dimerization interface in the VirB8 protein family.
Structure, 30, 2022
|
|
7JHP
 
 | |
4LD2
 
 | | Crystal structure of NE0047 in complex with cytidine | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Bitra, A, Biswas, A, Anand, R. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
4LCO
 
 | | Crystal structure of NE0047 with complex with substrate ammeline | | Descriptor: | 4,6-diamino-1,3,5-triazin-2-ol, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | Authors: | Bitra, A, Biswas, A, Anand, R. | | Deposit date: | 2013-06-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
4LCP
 
 | | Crytsal structure of NE0047 in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | Authors: | Bitra, A, Biswas, A, Anand, R. | | Deposit date: | 2013-06-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
4LD4
 
 | | Crystal structure of NE0047 in complex with cytosine | | Descriptor: | 6-AMINOPYRIMIDIN-2(1H)-ONE, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | Authors: | Bitra, A, Biswas, A, Anand, R. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
4LCN
 
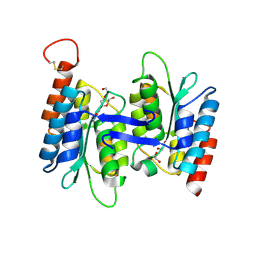 | | Crytsal structure of NE0047 in complex with 2'-DEOXY-GUANOSINE | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-GUANOSINE, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Anand, R, Bitra, A, Biswas, A. | | Deposit date: | 2013-06-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
5TLA
 
 | |
1UUT
 
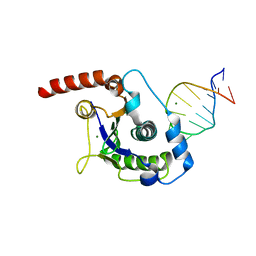 | | The Nuclease Domain of Adeno-Associated Virus Rep Complexed with the RBE' Stemloop of the Viral Inverted Terminal Repeat | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*TP*TP*GP *AP*GP*CP*TP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dyda, F, Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M. | | Deposit date: | 2004-01-10 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Nuclease Domain of Adeno-Associated Virus Rep Coordinates Replication Initiation Using Two Distinct DNA Recognition Interfaces
Mol.Cell, 13, 2004
|
|
2PWX
 
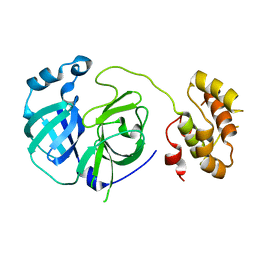 | | Crystal structure of G11A mutant of SARS-CoV 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Chen, S, Hu, T, Jiang, H, Shen, X. | | Deposit date: | 2007-05-14 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations.
J.Biol.Chem., 283, 2008
|
|
6WDR
 
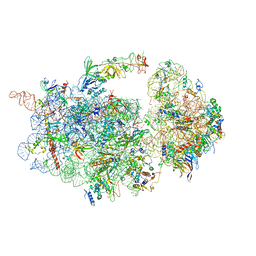 | | Subunit joining exposes nascent pre-40S rRNA for processing and quality control | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Rai, J, Parker, M.D, Huang, H, Choy, S, Ghalei, H, Johnson, M.C, Karbstein, K, Stroupe, M.E. | | Deposit date: | 2020-04-01 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An open interface in the pre-80S ribosome coordinated by ribosome assembly factors Tsr1 and Dim1 enables temporal regulation of Fap7.
Rna, 27, 2021
|
|
1A6C
 
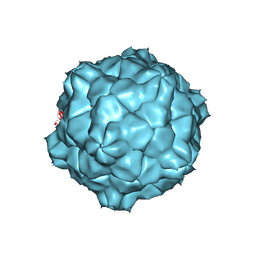 | | STRUCTURE OF TOBACCO RINGSPOT VIRUS | | Descriptor: | TOBACCO RINGSPOT VIRUS CAPSID PROTEIN | | Authors: | Johnson, J.E, Chandrasekar, V. | | Deposit date: | 1998-02-23 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of tobacco ringspot virus: a link in the evolution of icosahedral capsids in the picornavirus superfamily.
Structure, 6, 1998
|
|
1XGR
 
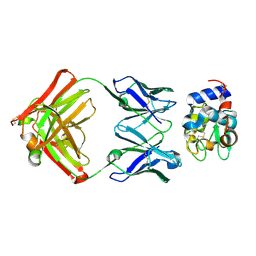 | | Structure for antibody HyHEL-63 Y33I mutant complexed with hen egg lysozyme | | Descriptor: | Lysozyme C, antibody kappa heavy chain, antibody kappa light chain | | Authors: | Li, Y, Huang, Y, Swaminathan, C.P, Smith-Gill, S.J, Mariuzza, R.A. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-06 | | Last modified: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces
Structure, 13, 2005
|
|
1XGU
 
 | | Structure for antibody HyHEL-63 Y33F mutant complexed with hen egg lysozyme | | Descriptor: | Lysozyme C, antibody kappa heavy chain, antibody kappa light chain | | Authors: | Li, Y, Huang, Y, Swaminathan, C.P, Smith-Gill, S.J, Mariuzza, R.A. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-06 | | Last modified: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces
Structure, 13, 2005
|
|
4KZ1
 
 | |
