3OC0
 
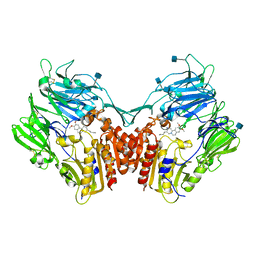 | | Structure of human DPP-IV with HTS hit (2S,3S,11bS)-3-butyl-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-ylamine | | Descriptor: | (2S,3R,11bR)-3-butyl-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6W2T
 
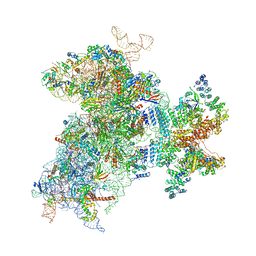 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the closed state (Class 2) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
1VBZ
 
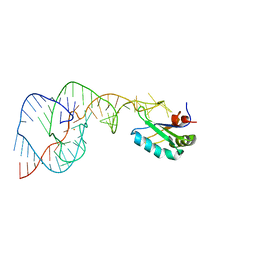 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Ba2+ solution | | Descriptor: | BARIUM ION, Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
3KJQ
 
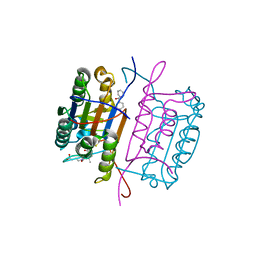 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
5CJQ
 
 | |
7JNH
 
 | | Crystal structure of a double-ENE RNA stability element in complex with a 28-mer poly(A) RNA | | Descriptor: | 28-mer poly(A) RNA, COBALT HEXAMMINE(III), Core double ENE RNA (Xtal construct) from Oryza sativa transposon,Core double ENE RNA (Xtal construct) from Oryza sativa transposon, ... | | Authors: | Torabi, S.F, Vaidya, A.T, Tycowski, K.T, DeGregorio, S.J, Wang, J, Shu, M.D, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | RNA stabilization by a poly(A) tail 3'-end binding pocket and other modes of poly(A)-RNA interaction.
Science, 371, 2021
|
|
1O5W
 
 | | The structure basis of specific recognitions for substrates and inhibitors of rat monoamine oxidase A | | Descriptor: | Amine oxidase [flavin-containing] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | Ma, J, Yoshimura, M, Yamashita, E, Nakagawa, A, Ito, A, Tsukihara, T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of rat monoamine oxidase a and its specific recognitions for substrates and inhibitors.
J.Mol.Biol., 338, 2004
|
|
1JWY
 
 | | CRYSTAL STRUCTURE OF THE DYNAMIN A GTPASE DOMAIN COMPLEXED WITH GDP, DETERMINED AS MYOSIN FUSION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Niemann, H.H, Knetsch, M.L.W, Scherer, A, Manstein, D.J, Kull, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms.
EMBO J., 20, 2001
|
|
5SUU
 
 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17-36. X-ray diffractometer data set. (ORN)CVFFCED(ORN)AII(SAR)L(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-PHE-CYS-GLU-ASP-ORN-ALA-ILE-ILE-SAR-LEU-ORN-VAL, CHLORIDE ION, IODIDE ION | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
7BCM
 
 | | The DDR1 Kinase Domain Bound To SR302 | | Descriptor: | Epithelial discoidin domain-containing receptor 1, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Roehm, S, Joerger, A, Knapp, S. | | Deposit date: | 2020-12-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
5TK9
 
 | | Structure of the HD-domain phosphohydrolase OxsA with Oxetanocin-A bound | | Descriptor: | MAGNESIUM ION, OxsA protein, [(2S,3R,4R)-4-(6-amino-9H-purin-9-yl)oxetane-2,3-diyl]dimethanol | | Authors: | Bridwell-Rabb, J, Drennan, C.L. | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1AV2
 
 | | Gramicidin A/CsCl complex, active as a dimer | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Burkhart, B.M, Li, N, Langs, D.A, Duax, W.L. | | Deposit date: | 1997-09-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Conducting Form of Gramicidin a is a Right-Handed Double-Stranded Double Helix.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2C1I
 
 | | Structure of Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) D 275 N Mutant. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PEPTIDOGLYCAN GLCNAC DEACETYLASE, SULFATE ION, ... | | Authors: | Blair, D.E, Schuttelkopf, A.W, MacRae, J.I, van Aalten, D.M.F. | | Deposit date: | 2005-09-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
5UGH
 
 | | Crystal structure of Mat2a bound to the allosteric inhibitor PF-02929366 | | Descriptor: | 2-(7-chloro-5-phenyl[1,2,4]triazolo[4,3-a]quinolin-1-yl)-N,N-dimethylethan-1-amine, S-adenosylmethionine synthase isoform type-2 | | Authors: | Kaiser, S.E, Feng, J, Stewart, A.E. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A.
Nat. Chem. Biol., 13, 2017
|
|
3B8D
 
 | | Fructose 1,6-bisphosphate aldolase from rabbit muscle | | Descriptor: | Fructose-bisphosphate aldolase A, SULFATE ION | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2007-11-01 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
1VBY
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, and Mn2+ bound | | Descriptor: | Hepatitis Delta virus ribozyme, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
3QUE
 
 | | Human p38 MAP Kinase in Complex with Skepinone-L | | Descriptor: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Mayer-Wrangowski, S, Richters, A, Rauh, D. | | Deposit date: | 2011-02-23 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor.
Nat.Chem.Biol., 8, 2012
|
|
5C9O
 
 | | Crystal structure of recombinant PLL lectin from Photorhabdus luminescens at 1.5 A resolution | | Descriptor: | GLYCEROL, PLL lectin | | Authors: | Kumar, A, Sykorova, P, Demo, G, Dobes, P, Hyrsl, P, Wimmerova, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed beta-Propeller Tetrameric Structure.
J.Biol.Chem., 291, 2016
|
|
1N6Z
 
 | |
2XZU
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND AN OXIDIZED PYRIMIDINE CONTAINING DNA AT 310K | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*VETP*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*G*A)-3', FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE, ... | | Authors: | Lebihan, Y.V, Izquierdo, M.A, Coste, F, Culard, F, Gehrke, T.H, Essalhi, K, Aller, P, Carrel, T, Castaing, B. | | Deposit date: | 2010-11-29 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 5-Hydroxy-5-Methylhydantoin DNA Lesion, a Molecular Trap for DNA Glycosylases
Nucleic Acids Res., 39, 2011
|
|
3AAZ
 
 | |
3H2K
 
 | | Crystal structure of a ligand-bound form of the rice cell wall degrading esterase LipA from Xanthomonas oryzae | | Descriptor: | esterase, octyl beta-D-glucopyranoside | | Authors: | Aparna, G, Chatterjee, A, Sonti, R.V, Sankaranarayanan, R. | | Deposit date: | 2009-04-14 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Cell Wall-Degrading Esterase of Xanthomonas oryzae Requires a Unique Substrate Recognition Module for Pathogenesis on Rice
Plant Cell, 21, 2009
|
|
2BM4
 
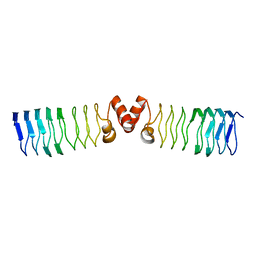 | | The Structure of MfpA (Rv3361c, C2 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
4QVH
 
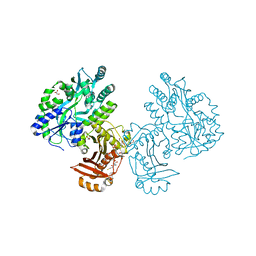 | | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein | | Descriptor: | CITRATE ANION, COENZYME A, GLYCEROL, ... | | Authors: | Jung, J, Bashiri, G, Johnston, J.M, Baker, E.N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein.
J.Struct.Biol., 188, 2014
|
|
1XKK
 
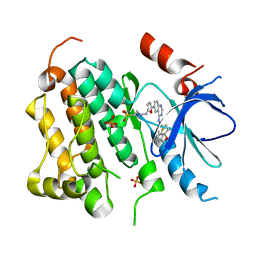 | | EGFR kinase domain complexed with a quinazoline inhibitor- GW572016 | | Descriptor: | Epidermal growth factor receptor, N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE, PHOSPHATE ION | | Authors: | Wood, E.R, Truesdale, A.T, McDonald, O.B, Yuan, D, Hassell, A, Dickerson, S.H, Ellis, B, Pennisi, C, Horne, E, Lackey, K, Alligood, K.J, Rusnak, D.W, Gilmer, T.M, Shewchuk, L.M. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells.
Cancer Res., 64, 2004
|
|
