8VZ3
 
 | |
8VZ5
 
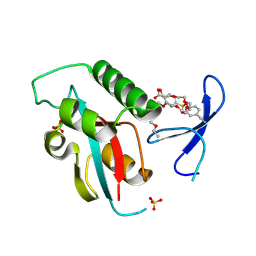 | | Structure of human PIN1 covalently derivatized with SuFEx compound | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-[fluoro(dihydroxy)-lambda~4~-sulfanyl]benzoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Louie, G.V, Noel, J.P, Wang, W.-M, Kelly, J.W. | | Deposit date: | 2024-02-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human PIN1 covalently derivatized with SuFEx compound
To be published
|
|
8VUM
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
8VQH
 
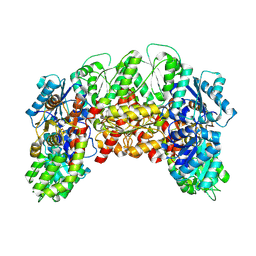 | |
8VIB
 
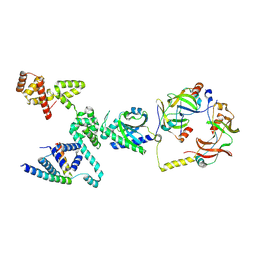 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VU9
 
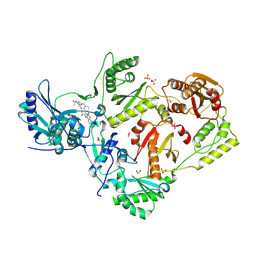 | | Crystal structure of wild-type HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5i3 | | Descriptor: | (2E)-3-[4-({2-[(1-{[4-(methanesulfonyl)phenyl]methyl}piperidin-4-yl)amino]pyrido[2,3-d]pyrimidin-4-yl}oxy)-3,5-dimethylphenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
8VUB
 
 | | Crystal structure of wild-type HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
8VYS
 
 | | Cryo-EM Structure of the BRAF V600E monomer bound to PLX8394 | | Descriptor: | (3S)-N-{3-[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl}-3-fluoropyrrolidine-1-sulfonamide, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
8VPK
 
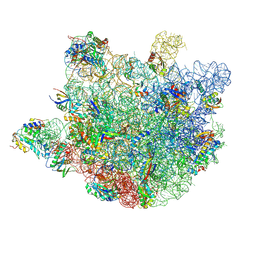 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX and erythromycin:50S-HflX-B-Ery | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L18, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-16 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
4MIY
 
 | | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | Deposit date: | 2013-09-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of myo-inositol dehydrogenase from Lactobacillus casei in complex with NAD and myo-inositol
To be Published
|
|
8VUF
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5i3 | | Descriptor: | (2E)-3-[4-({2-[(1-{[4-(methanesulfonyl)phenyl]methyl}piperidin-4-yl)amino]pyrido[2,3-d]pyrimidin-4-yl}oxy)-3,5-dimethylphenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
8VYP
 
 | | Cryo-EM Structure of the BRAF V600E monomer | | Descriptor: | 14-3-3 protein zeta/delta, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
8VYO
 
 | | Cryo-EM Structure of the BRAF WT monomer | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf, ZINC ION | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
8VYU
 
 | | Cryo-EM Structure of the BRAF WT monomer bound to PLX8394 | | Descriptor: | (3S)-N-{3-[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl}-3-fluoropyrrolidine-1-sulfonamide, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
8VYW
 
 | | Cryo-EM Structure of the BRAF D594G monomer | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
8VYV
 
 | | Cryo-EM Structure of the BRAF K601E monomer | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (5.86 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
6SNU
 
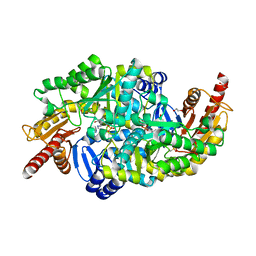 | | Crystal structure of the W60C mutant of the (S)-selective transaminase from Chromobacterium violaceum | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ruggieri, F, Gustafsson, C, Kimbung, R.Y, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-08-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Combined with Molecular Dynamics Reveal Altered Flow of Water in the Active Site of W60C Chromobacterium violaceum omega-transaminase
Not Published
|
|
8VTE
 
 | | Co-structure of the Fab of the anti-TIGIT Vibostolimab antibody with its antigen | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, Tiragolumab antibody heavy chain variable domains, Tiragolumab antibody light chain variable domains | | Authors: | Fischmann, T, Bahmanjah, S. | | Deposit date: | 2024-01-26 | | Release date: | 2025-05-28 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pharmacological and structural characterization of vibostolimab, a novel anti-TIGIT blocking antibody for cancer immunotherapy.
J Immunother Cancer, 13, 2025
|
|
8VYQ
 
 | | Cryo-EM Structure of the BRAF V600K monomer | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
4MKA
 
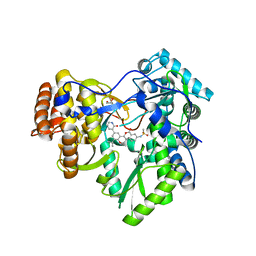 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 13 (N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{3-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1-oxo-1H-isochromen-7-yl}methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
8VTD
 
 | | Co-structure of the Fab of the anti-TIGIT Vibostolimab antibody with its antigen | | Descriptor: | GLYCEROL, T-cell immunoreceptor with Ig and ITIM domains, Vibostolimab Fab Heavy chain, ... | | Authors: | Fischmann, T. | | Deposit date: | 2024-01-26 | | Release date: | 2025-07-02 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Pharmacological and structural characterization of vibostolimab, a novel anti-TIGIT blocking antibody for cancer immunotherapy.
J Immunother Cancer, 13, 2025
|
|
1DEO
 
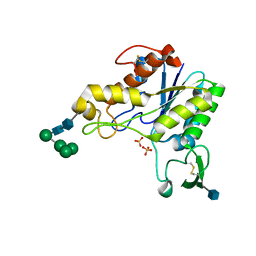 | | RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.55 A RESOLUTION WITH SO4 IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RHAMNOGALACTURONAN ACETYLESTERASE, SULFATE ION, ... | | Authors: | Molgaard, A, Kauppinen, S, Larsen, S. | | Deposit date: | 1999-11-15 | | Release date: | 2000-04-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases.
Structure Fold.Des., 8, 2000
|
|
8VYR
 
 | | Cryo-EM Structure of the BRAF V600E monomer bound to GDC0879 | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
1DED
 
 | | CRYSTAL STRUCTURE OF ALKALOPHILIC ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE COMPLEXED WITH AN INHIBITOR, ACARBOSE, AT 2.0 A RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-11-14 | | Release date: | 2000-04-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alkalophilic asparagine 233-replaced cyclodextrin glucanotransferase complexed with an inhibitor, acarbose, at 2.0 A resolution.
J.Biochem.(Tokyo), 127, 2000
|
|
8VWE
 
 | | HIV-1 neutralizing antibody D5_AR bound to HIV-1 gp41 coiled-coil pocket IQN17 | | Descriptor: | Neutralizing antibody D5_AR Heavy Chain, Neutralizing antibody D5_AR Light Chain, Transmembrane protein gp41 IQN17 peptide | | Authors: | Filsinger Interrante, M.V, Tang, S, Fernandez, D, Kim, P.S. | | Deposit date: | 2024-02-01 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Utilizing Machine Learning to Improve Neutralization Potency of an HIV-1 Antibody Targeting the gp41 N-Heptad Repeat.
Acs Chem.Biol., 20, 2025
|
|
