3PVY
 
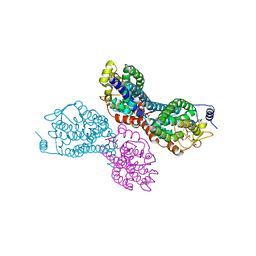 | |
4LXF
 
 | | Crystal structure of M. tuberculosis TreS | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Roy, R, Besra, G.S, Futterer, K. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis of alpha-glucan in mycobacteria involves a hetero-octameric complex of trehalose synthase TreS and Maltokinase Pep2.
Acs Chem.Biol., 8, 2013
|
|
3AES
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3QDS
 
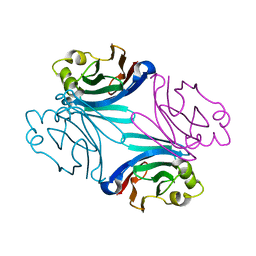 | | Structure of apo Boletus edulis lectin | | Descriptor: | BOLETUS EDULIS LECTIN | | Authors: | Bovi, M, Carrizo, M.E, Capaldi, S, Perduca, M, Chiarelli, L.R, Galliano, M, Monaco, H.L. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of a lectin with antitumoral properties in king bolete (Boletus edulis) mushrooms.
Glycobiology, 21, 2011
|
|
3QDX
 
 | | Structure of the orthorhombic form of the Boletus edulis lectin in complex with T-antigen disaccharide and N,N-diacetyl chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BOLETUS EDULIS LECTIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Bovi, M, Carrizo, M.E, Capaldi, S, Perduca, M, Chiarelli, L.R, Galliano, M, Monaco, H.L. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a lectin with antitumoral properties in king bolete (Boletus edulis) mushrooms.
Glycobiology, 21, 2011
|
|
4PLS
 
 | | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure. | | Descriptor: | ACETATE ION, Arm00010, CALCIUM ION | | Authors: | Reichen, C, Madhurantakam, C, Plueckthun, A, Mittl, P.R. | | Deposit date: | 2014-05-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure.
Protein Sci., 23, 2014
|
|
3QS1
 
 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
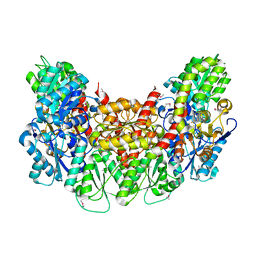
4PFK
 
 | | PHOSPHOFRUCTOKINASE. STRUCTURE AND CONTROL | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Evans, P.R, Hudson, P.J. | | Deposit date: | 1988-01-25 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphofructokinase: structure and control.
Philos.Trans.R.Soc.London,Ser.B, 293, 1981
|
|
3QDY
 
 | | Structure of the hexagonal form of the Boletus edulis lectin in complex with T-antigen disaccharide and N,N-diacetyl chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BOLETUS EDULIS LECTIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Bovi, M, Carrizo, M.E, Capaldi, S, Perduca, M, Chiarelli, L.R, Galliano, M, Monaco, H.L. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a lectin with antitumoral properties in king bolete (Boletus edulis) mushrooms.
Glycobiology, 21, 2011
|
|
3CL6
 
 | | Crystal structure of Puue Allantoinase | | Descriptor: | Puue allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
4PLO
 
 | |
3QKW
 
 | | Structure of Streptococcus parasangunini Gtf3 glycosyltransferase | | Descriptor: | Nucleotide sugar synthetase-like protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, F, Erlandsen, H, Huang, Y, Ding, L, Zhou, M, Liang, X, Ma, J.-B, Wu, H. | | Deposit date: | 2011-02-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Analysis of a New Subfamily of Glycosyltransferases Required for Glycosylation of Serine-rich Streptococcal Adhesins.
J.Biol.Chem., 286, 2011
|
|
4PV3
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, SODIUM ION | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
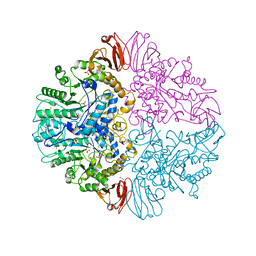
3AB4
 
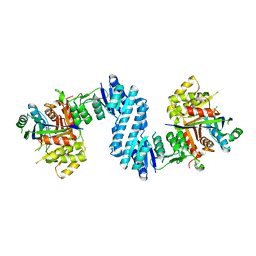 | | Crystal structure of feedback inhibition resistant mutant of aspartate kinase from Corynebacterium glutamicum in complex with lysine and threonine | | Descriptor: | Aspartokinase, LYSINE, THREONINE | | Authors: | Yoshida, A, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-11-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Mechanism of concerted inhibition of {alpha}2{beta}2-type heterooligomeric aspartate kinase from Corynebacterium glutamicum
J.Biol.Chem., 285, 2010
|
|
1E7W
 
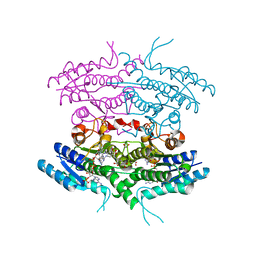 | | One active site, two modes of reduction correlate the mechanism of leishmania pteridine reductase with pterin metabolism and antifolate drug resistance in trpanosomes | | Descriptor: | 1,2-ETHANEDIOL, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gourley, D.G, Hunter, W.N. | | Deposit date: | 2000-09-11 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pteridine Reductase Mechanism Correlates Pterin Metabolism with Drug Resistance in Trypanosomatid Parasites.
Nat.Struct.Biol., 8, 2001
|
|
1EIA
 
 | |
3QRW
 
 | |
3QDV
 
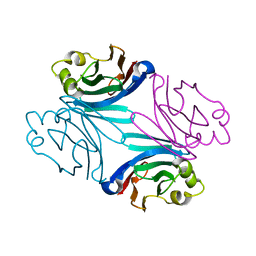 | | Structure of the orthorhombic form of the Boletus edulis lectin in complex with N-acetyl glucosamine and N-acetyl galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-alpha-D-glucopyranose, BOLETUS EDULIS LECTIN | | Authors: | Bovi, M, Carrizo, M.E, Capaldi, S, Perduca, M, Chiarelli, L.R, Galliano, M, Monaco, H.L. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a lectin with antitumoral properties in king bolete (Boletus edulis) mushrooms.
Glycobiology, 21, 2011
|
|
7SEH
 
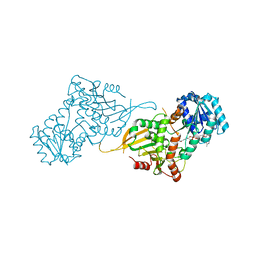 | | Glucose-6-phosphate 1-dehydrogenase (K403QdLtL) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
1ECG
 
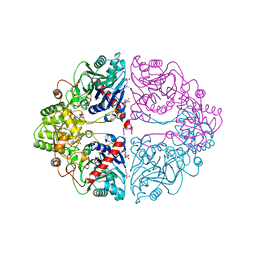 | | DON INACTIVATED ESCHERICHIA COLI GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE (PRPP) AMIDOTRANSFERASE | | Descriptor: | 5-OXO-L-NORLEUCINE, GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE AMIDOTRANSFERASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Krahn, J.M. | | Deposit date: | 1996-04-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site.
J.Biol.Chem., 271, 1996
|
|
3IOL
 
 | |
8FZF
 
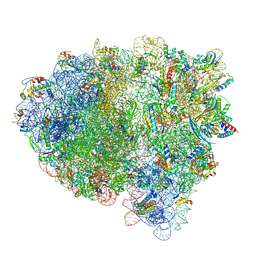 | | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP
Nat.Struct.Mol.Biol., 2024
|
|
3SPJ
 
 | |
3SPG
 
 | | Inward rectifier potassium channel Kir2.2 R186A mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
7N53
 
 | | Complex structure of PAP4 with papaverine | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, GLYCEROL, PAP4, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
