6S2A
 
 | | PI PLC mutant H82A | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase | | Authors: | Eisenreichova, A, Boura, E. | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PI PLC mutant H82A
To Be Published
|
|
5N9N
 
 | | Crystal structure of human Protein kinase CK2 catalytic subunit in complex with the ATP-competitive, tight-binding dibenzofuran inhibitor TF85 (4a) | | Descriptor: | (4~{Z})-7,9-bis(chloranyl)-4-[[(4-methoxyphenyl)amino]methylidene]-8-oxidanyl-1,2-dihydrodibenzofuran-3-one, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Schnitzler, A, Gratz, A, Bollacke, A, Weyrich, M, Kucklaender, U, Wuensch, B, Goetz, C, Niefind, K, Jose, J. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | A pi-Halogen Bond of Dibenzofuranones with the Gatekeeper Phe113 in Human Protein Kinase CK2 Leads to Potent Tight Binding Inhibitors.
Pharmaceuticals, 11, 2018
|
|
9KE9
 
 | | Crystal structure of the PIN1 and fragment 22 complex. | | Descriptor: | (2H-1,3-benzodioxol-5-yl)methanol, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-04 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9KEW
 
 | | Crystal structure of the PIN1 and fragment 29 complex. | | Descriptor: | 2,3-dihydro-1-benzofuran-7-ylmethanol, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9KEY
 
 | | Crystal structure of the PIN1 and fragment 30 complex. | | Descriptor: | 2,1,3-benzothiadiazol-5-ylmethanol, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9KEZ
 
 | | Crystal structure of the PIN1 and fragment 31 complex. | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-5-ylmethanol, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9KXI
 
 | | Crystal structure of the PIN1 and fragment 31 complex. | | Descriptor: | 2-(2-methylimidazol-1-yl)aniline, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-12-06 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9F2F
 
 | | Carbonic anhydrase II variant with bound iron complex in space group R3 (ArPase) | | Descriptor: | 1,2-ETHANEDIOL, 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, Carbonic anhydrase 2, ... | | Authors: | Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-23 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
6G4O
 
 | | Non-aged form of Torpedo californica acetylcholinesterase inhibited by tabun analog NEDPA bound to uncharged reactivator 1 | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
9F2E
 
 | | Carbonic anhydrase II variant with bound iron complex in space group C2 (ArPase) | | Descriptor: | 1,2-ETHANEDIOL, 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, Carbonic anhydrase 2, ... | | Authors: | Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-23 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
9KPT
 
 | |
3LV8
 
 | | 1.8 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with TMP, thymidine-5'-diphosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with TMP, thymidine-5'-diphosphate and ADP
To be Published
|
|
4YK6
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A4 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
5O5A
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ032 | | Descriptor: | 1-ethyl-~{N}-methyl-2,3-bis(oxidanylidene)-~{N}-[(1-phenylpyrazol-4-yl)methyl]-4~{H}-quinoxaline-6-carboxamide, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5LBY
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with crenolanib | | Descriptor: | 1-(2-{5-[(3-Methyloxetan-3-yl)methoxy]-1H-benzimidazol-1-yl}quinolin-8-yl)piperidin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuester, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
7MQP
 
 | | E. coli dihydrofolate reductase complexed with 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-4'-methyl[1,1'-biphenyl]-4-carboxamide, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
8PF5
 
 | | Crystal structure of Trypanosoma brucei trypanothione reductase in complex with 1-(3,4-dichlorobenzyl)-4-(((5-((4-fluorophenethyl)carbamoyl)furan-2-yl)methyl)carbamoyl)-1-(3-phenylpropyl)piperazin-1-ium | | Descriptor: | 4-[(3,4-dichlorophenyl)methyl]-~{N}-[[5-[2-(4-fluorophenyl)ethylcarbamoyl]furan-2-yl]methyl]-4-(3-phenylpropyl)-1,4$l^{4}-diazinane-1-carboxamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Exertier, C, Antonelli, L, Fiorillo, A, Ilari, A. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fragment Merging, Growing, and Linking Identify New Trypanothione Reductase Inhibitors for Leishmaniasis.
J.Med.Chem., 67, 2024
|
|
9KSH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-pyridin-3-yl-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
7U4K
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
9M3R
 
 | |
6E4Q
 
 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9A in Complex with UDP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
9D8E
 
 | | Crystal structure of the ACVR1 (ALK2) Kinase Domain in complex with inhibitor CDD-2789 | | Descriptor: | 1-cyclobutyl-N-[3-(dimethylamino)propyl]-2-(3,4,5-trimethoxyphenyl)-1H-1,3-benzimidazole-6-carboxamide, Activin receptor type-1, GLYCEROL, ... | | Authors: | Ta, H.M, Kim, C, Jimmidi, K, Matzuk, M.M. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of highly potent and ALK2/ALK1 selective kinase inhibitors using DNA-encoded chemistry technology.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7LJ1
 
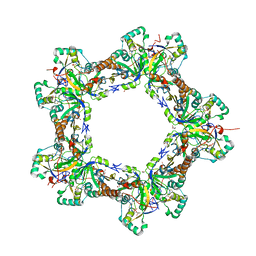 | | Human Prx1-Srx Decameric Complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Forshaw, T.E, Reisz, J.A, Nelson, K.J, Gumpena, R, Lawson, J.R, Jonsson, T, Wu, H, Clodfelter, J.E, Johnson, L, Furdui, C.M, Lowther, W.T. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Specificity of Human Sulfiredoxin for Reductant and Peroxiredoxin Oligomeric State.
Antioxidants (Basel), 10, 2021
|
|
5DCW
 
 | | Iridoid synthase from Catharanthus roseus - ligand free structure | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
6C4N
 
 | | Pseudopaline dehydrogenase (PaODH) - NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
