8BCW
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
6FNK
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with a pyrazolo[3,4-d]pyrimidine fragment of NVP-BHG712 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-amine, Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.049 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
6HMJ
 
 | | Structure of an RNA-binding Light-Oxygen-Voltage Receptor | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Ziegler, T, Moniot, S, Moeglich, A. | | Deposit date: | 2018-09-12 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | A blue light receptor that mediates RNA binding and translational regulation.
Nat.Chem.Biol., 15, 2019
|
|
5YFM
 
 | |
6IN0
 
 | | Crystal structure of EphA3 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Ephrin type-A receptor 3 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
4Z08
 
 | |
6RJC
 
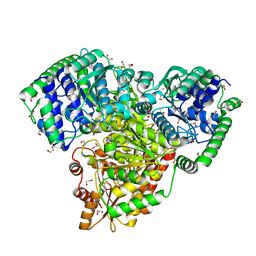 | |
7S3D
 
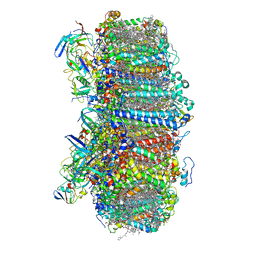 | | Structure of photosystem I with bound ferredoxin from Synechococcus sp. PCC 7335 acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2Fe-2S ferredoxin-type domain-containing protein, ... | | Authors: | Gisriel, C.J, Flesher, D.A, Shen, G, Wang, J, Ho, M, Brudvig, G.W, Bryant, D.A. | | Deposit date: | 2021-09-05 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure of a photosystem I-ferredoxin complex from a marine cyanobacterium provides insights into far-red light photoacclimation.
J.Biol.Chem., 298, 2021
|
|
7UGR
 
 | | Crystal structure of hyperfolder YFP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyperfolder yellow fluorescent protein, ... | | Authors: | Campbell, B.C, Liu, C.F, Petsko, G.A. | | Deposit date: | 2022-03-25 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chemically stable fluorescent proteins for advanced microscopy.
Nat.Methods, 19, 2022
|
|
8PE2
 
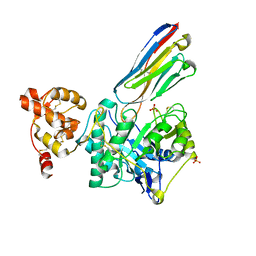 | | Crystal structure of Gel4 in complex with Nanobody 3 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 3, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7VBF
 
 | |
6H2R
 
 | |
8OTV
 
 | | Crystal structure of NUDT14 complexed with novel compound | | Descriptor: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Balikci, E, Feyerherm, C, Bradshaw, W, Seupel, R, Brennan, P.E, Bountra, C, von Delft, F, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
8BXX
 
 | | Crystal structure of formate dehydrogenase FDH2 enzyme from Granulicella mallensis MP5ACTX8 in complex with NAD and azide. | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, Formate dehydrogenase, ... | | Authors: | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | Deposit date: | 2022-12-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | From the amelioration of a NADP+-dependent formate dehydrogenase to the discovery of a new enzyme: round trip from theory to practice
ChemCatChem, 2020
|
|
4ZW0
 
 | |
7V39
 
 | | Crystal structure of NP exonuclease-PCMB complex | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Nucleoprotein, ZINC ION | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
4Y75
 
 | | Yeast 20S proteasome in complex with Ac-PAF-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PAF-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y81
 
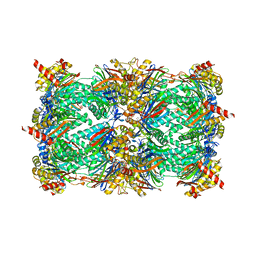 | | Yeast 20S proteasome in complex with Ac-PAY-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PAY-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
5Y4N
 
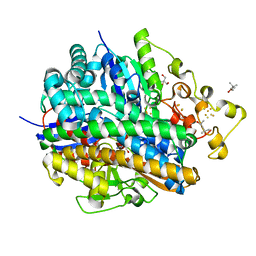 | | Crystal structure of aerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J.Inorg.Biochem., 177, 2017
|
|
7VC6
 
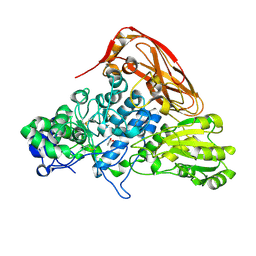 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, xylan 1,4-beta-xylosidase | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
8AUE
 
 | |
6IS2
 
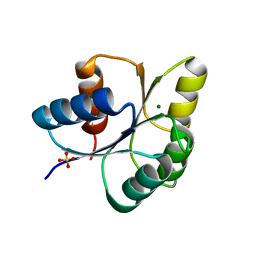 | |
6I4P
 
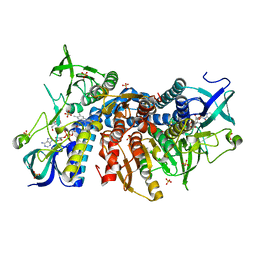 | | Crystal structure of the disease-causing G194C mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Szabo, E, Wilk, P, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
5M1I
 
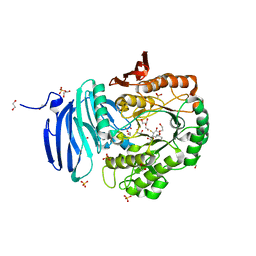 | | Structure of GH36 alpha-galactosidase from Thermotoga maritima in a covalent complex with a cyclopropyl carbasugar. | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},6~{R})-4-fluoranyl-1-(hydroxymethyl)bicyclo[4.1.0]heptane-2,3-diol, 1,2-ETHANEDIOL, Alpha-galactosidase, ... | | Authors: | Pengelly, R, Gloster, T. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Snapshots for Mechanism-Based Inactivation of a Glycoside Hydrolase by Cyclopropyl Carbasugars.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6I59
 
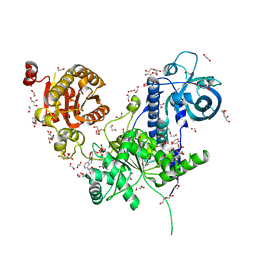 | | Long wavelength native-SAD phasing of Sen1 helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
