4ZHJ
 
 | |
8RUR
 
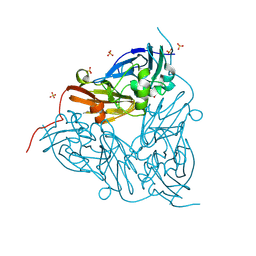 | | High pH (8.0) nitrite-bound MSOX movie series dataset 2 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [1.38 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8SVU
 
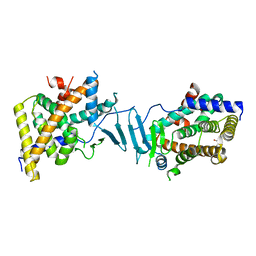 | | Crystal structure of the L428V mutant of pregnane X receptor ligand binding domain in apo form | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1 | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
9C0A
 
 | | Sigma class glutathione transferase from Taenia solium 1.75 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Miranda-Blancas, R, Cardona-Echavarria, M.C, Garcia-Gutierrez, P, Sanchez-Juarez, C, Flores-Lopez, R, Zubillaga, R, Rodriguez-Lima, O, Sanchez-Perez, L.C, Landa, A, Rudino-Pinera, E. | | Deposit date: | 2024-05-24 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into sigma class glutathione transferase from Taenia solium: Analysis and functional implications.
Plos Negl Trop Dis, 19, 2025
|
|
8TEC
 
 | |
6AAL
 
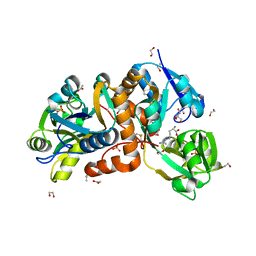 | | Crystal Structure of putative amino acid binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus in complex with Arginine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
5DY2
 
 | | Crystal structure of Dbr2 with mutation M27L | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
9LGV
 
 | | Crystal structure of human PKMYT1 protein kinase domain with Naphthyridinone Inhibitor compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-7-chloranyl-4-(7-fluoranyl-1H-indazol-4-yl)-1H-1,5-naphthyridin-2-one, L(+)-TARTARIC ACID, ... | | Authors: | Xu, Z.H, Chen, S. | | Deposit date: | 2025-01-10 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Discovery of Naphthyridinone Derivatives as Selective and Potent PKMYT1 Inhibitors with Antitumor Efficacy.
J.Med.Chem., 68, 2025
|
|
5NK2
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2b | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6GW2
 
 | |
5MZ2
 
 | | Rubisco from Thalassiosira antarctica | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Andersson, I, Valegard, K. | | Deposit date: | 2017-01-30 | | Release date: | 2018-02-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analyses of Rubisco from arctic diatom species reveal unusual posttranslational modifications.
J. Biol. Chem., 293, 2018
|
|
9FDR
 
 | | Crystal structure of human Sirt2 in apo form with opened selectivity pocket | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Friedrich, F, Einsle, O, Jung, M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Efficient Crystallization of Apo Sirt2 for Small-Molecule Soaking and Structural Analysis of Ligand Interactions.
J.Med.Chem., 68, 2025
|
|
9FRU
 
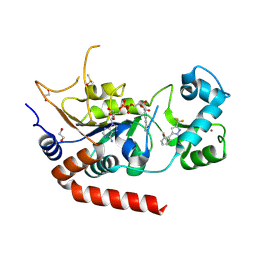 | | Crystal structure of human Sirt2 in complex with a pyrazole-based fragment inhibitor and NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Friedrich, F, Einsle, O, Jung, M. | | Deposit date: | 2024-06-19 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Efficient Crystallization of Apo Sirt2 for Small-Molecule Soaking and Structural Analysis of Ligand Interactions.
J.Med.Chem., 68, 2025
|
|
9FDU
 
 | | Crystal structure of human Sirt2 in complex with a pyridine-3-carbothioamide-based fragment inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-[2,2,2-tris(fluoranyl)ethoxy]pyridine-3-carbothioamide, CHLORIDE ION, ... | | Authors: | Friedrich, F, Einsle, O, Jung, M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Efficient Crystallization of Apo Sirt2 for Small-Molecule Soaking and Structural Analysis of Ligand Interactions.
J.Med.Chem., 68, 2025
|
|
9FDW
 
 | | Crystal structure of human Sirt2 in complex with a 3-chlorobenzamide-based fragment inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-chloranyl-~{N}-(pyridin-2-ylmethyl)benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Friedrich, F, Einsle, O, Jung, M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Efficient Crystallization of Apo Sirt2 for Small-Molecule Soaking and Structural Analysis of Ligand Interactions.
J.Med.Chem., 68, 2025
|
|
9FDT
 
 | | Crystal structure of human Sirt2 in complex with a pyrazole-based fragment inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Friedrich, F, Einsle, O, Jung, M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Efficient Crystallization of Apo Sirt2 for Small-Molecule Soaking and Structural Analysis of Ligand Interactions.
J.Med.Chem., 68, 2025
|
|
5CY4
 
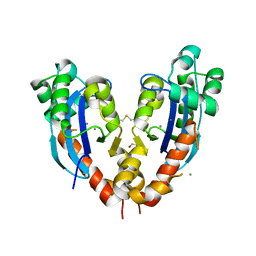 | |
5NKB
 
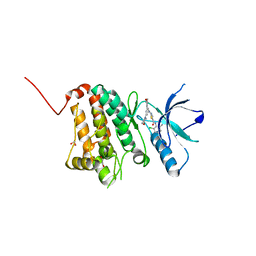 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 4a | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[(3,5-dimorpholin-4-ylphenyl)amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
7RB7
 
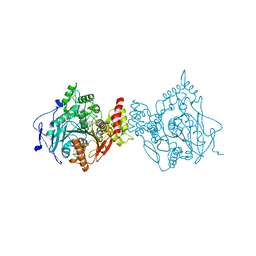 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA and MMB4 oxime | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
6HAR
 
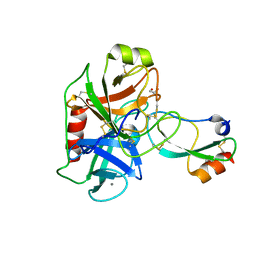 | | Crystal structure of Mesotrypsin in complex with APPI-M17C/I18F/F34C | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, CALCIUM ION, ... | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N. | | Deposit date: | 2018-08-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J.Biol.Chem., 294, 2019
|
|
9FNC
 
 | | Half-closed CODH/ACS in the carbonylated state | | Descriptor: | CARBON MONOXIDE, CO-dehydrogenase, CO-methylating acetyl-CoA synthase, ... | | Authors: | Ruickoldt, J, Wendler, P, Dobbek, H. | | Deposit date: | 2024-06-10 | | Release date: | 2025-06-25 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Ligand binding to a Ni-Fe cluster orchestrates conformational changes of the CO-dehydrogenase-acetyl-CoA synthase complex.
Nat Catal, 8, 2025
|
|
8HP3
 
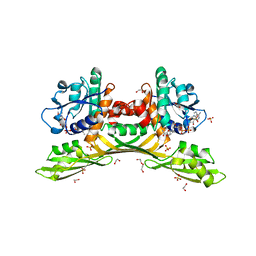 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
5N81
 
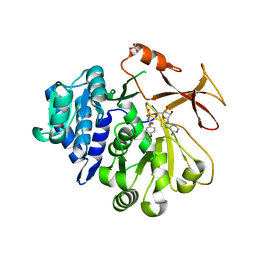 | | Crystal structure of an engineered TycA variant in complex with an O-propargyl-beta-Tyr-AMP analog | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Tyrocidine synthase 1, ... | | Authors: | Niquille, D.L, Hansen, D.A, Mori, T, Fercher, D, Kries, H, Hilvert, D. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonribosomal biosynthesis of backbone-modified peptides.
Nat Chem, 10, 2018
|
|
6H5T
 
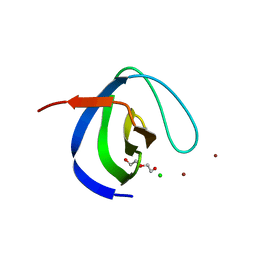 | | Intersectin SH3A short isoform | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Driller, J.H, Gerth, F, Freund, C, Wahl, M.C, Loll, B. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Exon Inclusion Modulates Conformational Plasticity and Autoinhibition of the Intersectin 1 SH3A Domain.
Structure, 27, 2019
|
|
5HZP
 
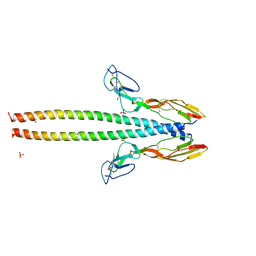 | | Structure of human C4b-binding protein alpha chain CCP domains 1 and 2 in complex with the hypervariable region of group A Streptococcus M49 protein. | | Descriptor: | C4b-binding protein alpha chain, M protein, serotype 49, ... | | Authors: | Buffalo, C.Z, Bahn-Suh, A.J, Ghosh, P. | | Deposit date: | 2016-02-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein.
Nat Microbiol, 1, 2016
|
|
