1KBA
 
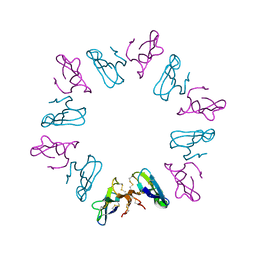 | |
4QOZ
 
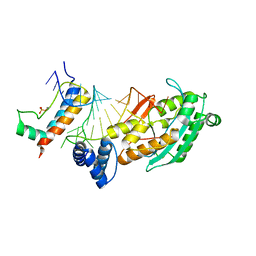 | |
4JPC
 
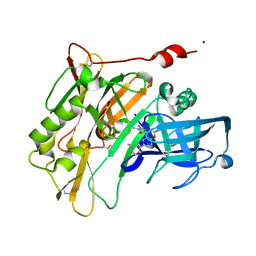 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | Descriptor: | 3-[(4R)-2'-amino-1',2,2-trimethyl-5'-oxo-1',2,3,5'-tetrahydrospiro[chromene-4,4'-imidazol]-6-yl]benzonitrile, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
1NCT
 
 | | TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
4J8Z
 
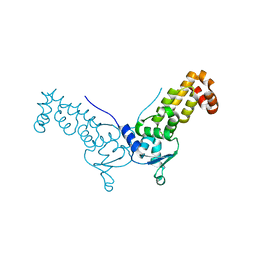 | |
1IA9
 
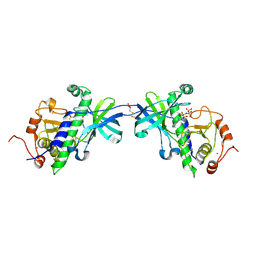 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (AMPPNP COMPLEX) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TRANSIENT RECEPTOR POTENTIAL-RELATED PROTEIN, ... | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
3ENL
 
 | |
4JP9
 
 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | Descriptor: | (4R)-2'-amino-6-(3-chlorophenyl)-1',2,2-trimethyl-2,3-dihydrospiro[chromene-4,4'-imidazol]-5'(1'H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
3CLB
 
 | | Structure of bifunctional TcDHFR-TS in complex with TMQ | | Descriptor: | 1,2-ETHANEDIOL, DHFR-TS, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3R1G
 
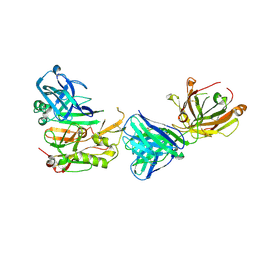 | | Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody | | Descriptor: | Beta-secretase 1, FAB of YW412.8.31 antibody heavy chain, FAB of YW412.8.31 antibody light chain | | Authors: | Wang, W, Rouge, L, Wu, P, Chiu, C, Chen, Y, Wu, Y, Watts, R.J. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-{beta} Production in Vivo.
Sci Transl Med, 3, 2011
|
|
1IFG
 
 | | CRYSTAL STRUCTURE OF A MONOMERIC FORM OF GENERAL PROTEASE INHIBITOR, ECOTIN IN ABSENCE OF A PROTEASE | | Descriptor: | ECOTIN | | Authors: | Eggers, C.T, Wang, S.X, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of ecotin dimerization in protease inhibition.
J.Mol.Biol., 308, 2001
|
|
2P32
 
 | |
1IIC
 
 | |
1J2Q
 
 | | 20S proteasome in complex with calpain-Inhibitor I from archaeoglobus fulgidus | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID [1-(1-FORMYL-PENTYLCARBAMOYL)-3-METHYL-BUTYL]-AMIDE, Proteasome alpha subunit, Proteasome beta subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
2LXP
 
 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2LXH
 
 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
1SEP
 
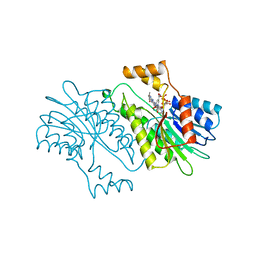 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND SEPIAPTERIN | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SEPIAPTERIN REDUCTASE | | Authors: | Auerbach, G, Herrmann, A, Guetlich, M, Fischer, M, Jacob, U, Bacher, A, Huber, R. | | Deposit date: | 1997-05-23 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
1SPI
 
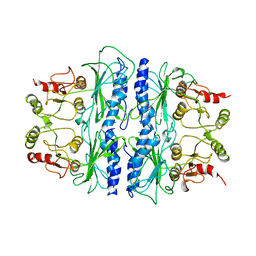 | | CRYSTAL STRUCTURE OF SPINACH CHLOROPLAST FRUCTOSE-1,6-BISPHOSPHATASE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Villeret, V, Huang, S, Zhang, Y, Xue, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-14 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of spinach chloroplast fructose-1,6-bisphosphatase at 2.8 A resolution.
Biochemistry, 34, 1995
|
|
6TIR
 
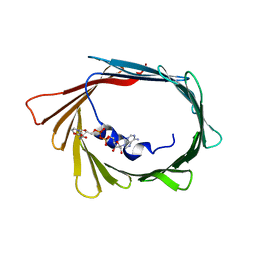 | |
1EQW
 
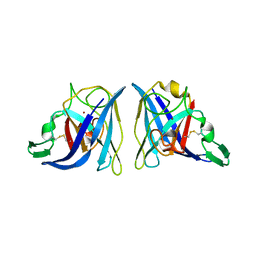 | | CRYSTAL STRUCTURE OF SALMONELLA TYPHIMURIUM CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Pesce, A, Battistoni, A, Stroppolo, M.E, Polizio, F, Nardini, M, Kroll, J.S, Langford, P.R, O'Neill, P, Sette, M, Desideri, A, Bolognesi, M. | | Deposit date: | 2000-04-06 | | Release date: | 2000-09-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene.
J.Mol.Biol., 302, 2000
|
|
6GX9
 
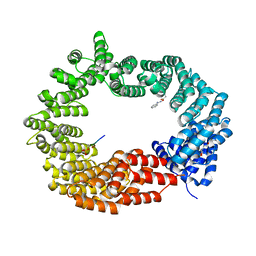 | | Crystal structure of the TNPO3 - CPSF6 RSLD complex | | Descriptor: | BENZAMIDINE, BICINE, Cleavage and polyadenylation specificity factor subunit 6, ... | | Authors: | Cherepanov, P, Cook, N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6.
Nucleic Acids Res., 47, 2019
|
|
6TU3
 
 | | Rat 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Deshmukh, F.K, Polkinghorn, C.R, Elad, N, Sharon, M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Comparative Structural Analysis of 20S Proteasome Ortholog Protein Complexes by Native Mass Spectrometry.
Acs Cent.Sci., 6, 2020
|
|
317D
 
 | |
5T19
 
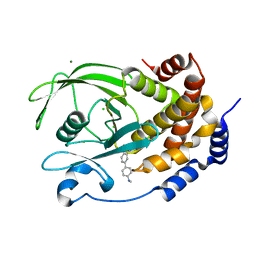 | | Structure of PTP1B complexed with N-(3'-(1,1-dioxido-4-oxo-1,2,5-thiadiazolidin-2-yl)-4'-methyl-[1,1'-biphenyl]-4-yl)acetamide | | Descriptor: | 5-[4-methyl-4'-(methylamino)[1,1'-biphenyl]-3-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Laciak, A.R, Tanner, J.J. | | Deposit date: | 2016-08-18 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | Covalent Allosteric Inactivation of Protein Tyrosine Phosphatase 1B (PTP1B) by an Inhibitor-Electrophile Conjugate.
Biochemistry, 56, 2017
|
|
344D
 
 | | DETERMINATION BY MAD-DM OF THE STRUCTURE OF THE DNA DUPLEX D(ACGTACG(5-BRU))2 AT 1.46A AND 100K | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*(BRU))-3') | | Authors: | Todd, A.R, Adams, A, Powell, H.R, Cardin, C.J. | | Deposit date: | 1997-08-04 | | Release date: | 1997-09-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Determination by MAD-DM of the structure of the DNA duplex d[ACGTACG(5-BrU)]2 at 1.46 A and 100 K.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
