7JL3
 
 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JMD
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JKC
 
 | | Sheep Connexin-46 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JLP
 
 | | cryo-EM structure of human ATG9A in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JMC
 
 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN1
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7K24
 
 | |
7K3V
 
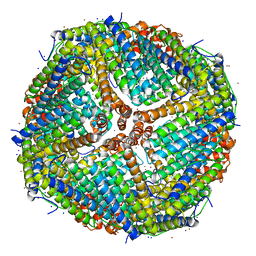 | | Apoferritin structure at 1.34 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with K3 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.34 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
7KTX
 
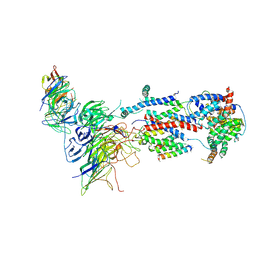 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to a Fab in DDM detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7KHE
 
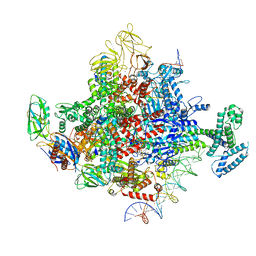 | | Escherichia coli RNA polymerase and rrnBP1 promoter pre-open complex with DksA/ppGpp | | Descriptor: | CHAPSO, DNA (46-MER), DNA (54-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
7KJR
 
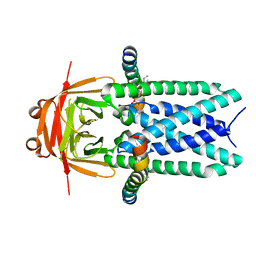 | | Cryo-EM structure of SARS-CoV-2 ORF3a | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Kern, D.M, Hoel, C.M, Kotecha, A, Brohawn, S.G. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7F5O
 
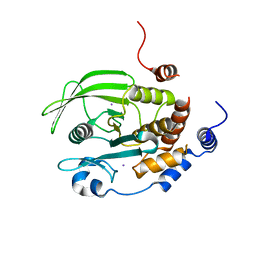 | | Crystal structure of PTPN2 catalytic domain | | Descriptor: | IODIDE ION, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Singh, J.P, Lin, M.-J, Hsu, S.-F, Lee, C.-C, Meng, T.-C. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of TCPTP Unravels an Allosteric Regulatory Role of Helix alpha 7 in Phosphatase Activity.
Biochemistry, 60, 2021
|
|
7EWA
 
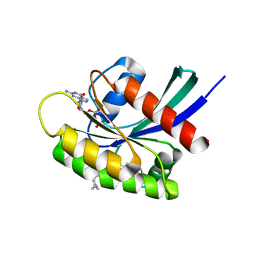 | | GDP-bound KRAS G12D in complex with TH-Z827 | | Descriptor: | 4-[(1~{R},5~{S})-3,8-diazabicyclo[3.2.1]octan-8-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
8AIF
 
 | |
7EU3
 
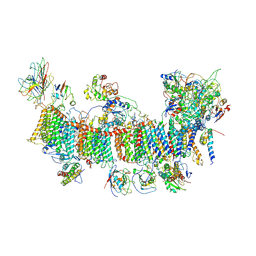 | | Chloroplast NDH complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EXP
 
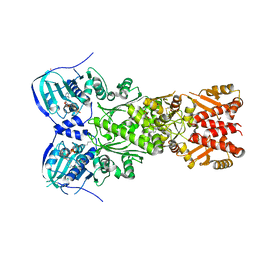 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | Descriptor: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7F6H
 
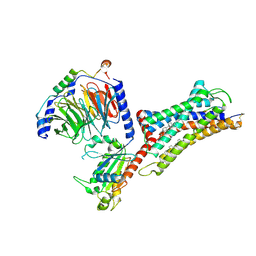 | | Cryo-EM structure of human bradykinin receptor BK2R in complex Gq proteins and bradykinin | | Descriptor: | Bradykinin, Bradykinin receptor BK2R, CHOLESTEROL, ... | | Authors: | Shen, J, Zhang, D, Fu, Y, Chen, A, Zhang, H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-01-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of human bradykinin receptor-G q proteins complexes.
Nat Commun, 13, 2022
|
|
7FHJ
 
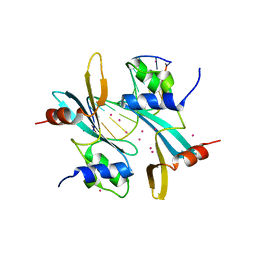 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
7FDE
 
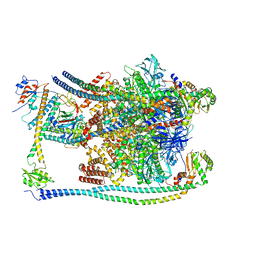 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | Descriptor: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7F8E
 
 | |
7F6I
 
 | | Cryo-EM structure of human bradykinin receptor BK2R in complex Gq proteins and kallidin | | Descriptor: | Bradykinin receptor BK2R, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, J, Zhang, D, Fu, Y, Chen, A, Zhang, H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-01-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human bradykinin receptor-G q proteins complexes.
Nat Commun, 13, 2022
|
|
7F3M
 
 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
8AD9
 
 | | Crystal structure of ClpC2 C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cyclomarin A, ... | | Authors: | Taylor, G, Cui, H.J, Leodolter, J, Giese, C, Weber-Ban, E. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | ClpC2 protects mycobacteria against a natural antibiotic targeting ClpC1-dependent protein degradation.
Commun Biol, 6, 2023
|
|
7EWK
 
 | | Barley photosystem I-LHCI-Lhca6 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
