6JNH
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferasefrom Acinetobacter baumannii with Ascorbic acid (Vitamin-C) at 2.0A resolution | | Descriptor: | ASCORBIC ACID, Phosphopantetheine adenylyltransferase, SODIUM ION, ... | | Authors: | Iqbal, N, Gupta, A, Sharm, P, Singh, T.P. | | Deposit date: | 2019-03-14 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Ascorbic acid (Vitamin-C) at 2.0A resolution
To Be Published
|
|
5N2E
 
 | | Structure of the E9 DNA polymerase from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
6JJS
 
 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | Descriptor: | ACETATE ION, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
5JRF
 
 | | Crystal structure of the light-driven sodium pump KR2 bound with iodide ions | | Descriptor: | EICOSANE, IODIDE ION, Sodium pumping rhodopsin | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5U99
 
 | | Transition state analysis of adenosine triphosphate phosphoribosyltransferase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Moggre, G.-J, Poulin, M.B, Tyler, P.C, Schramm, V.L, Parker, E.J. | | Deposit date: | 2016-12-15 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transition State Analysis of Adenosine Triphosphate Phosphoribosyltransferase.
ACS Chem. Biol., 12, 2017
|
|
5N0O
 
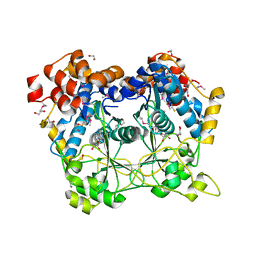 | |
6JGO
 
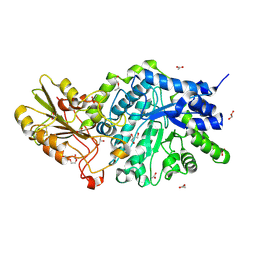 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JHA
 
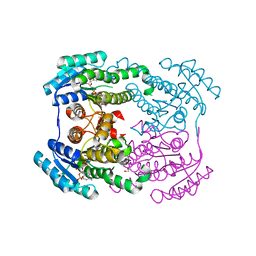 | | Crystal structure of NADPH bound AerF from Microcystis aeruginosa | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase family protein, TRIETHYLENE GLYCOL | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
5N1T
 
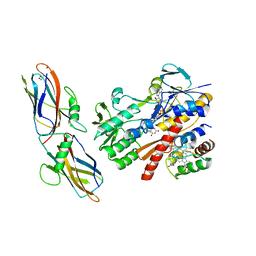 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | Descriptor: | COPPER (II) ION, CopC, Cytochrome C, ... | | Authors: | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5JY5
 
 | | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Bravo-Chaucanes, C.P, Abadio, A.K.R, Kioshima, E.S, Felipe, M.S.S, Barbosa, J.A.R.G. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution
To Be Published
|
|
6JIW
 
 | | Crystal structure of S. aureus CntK with C72S mutation | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Luo, S, Ju, Y, Zhou, H. | | Deposit date: | 2019-02-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus.
Int.J.Biol.Macromol., 135, 2019
|
|
5UGR
 
 | | Malyl-CoA lyase from Methylobacterium extorquens | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Malyl-CoA lyase/beta-methylmalyl-CoA lyase, ... | | Authors: | Gonzalez, J.M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Methylobacterium extorquens malyl-CoA lyase: CoA-substrate binding correlates with domain shift.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2WUE
 
 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPODA | | Descriptor: | (3E,5R)-8-(2-CHLOROPHENYL)-5-METHYL-2,6-DIOXOOCT-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, THIOCYANATE ION | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
5UHZ
 
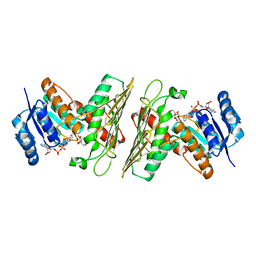 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium | | Descriptor: | (3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-one, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium
To be published
|
|
5UD6
 
 | |
6JJT
 
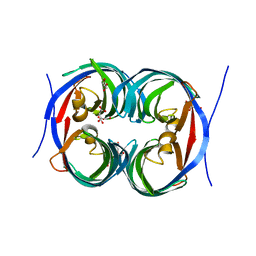 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
5JVW
 
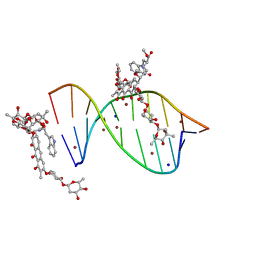 | | Crystal structure of mithramycin analogue MTM SA-Trp in complex with a 10-mer DNA AGAGGCCTCT. | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*GP*CP*CP*TP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Trp, ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2016-05-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
1DS8
 
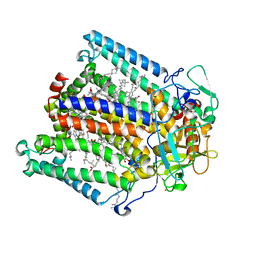 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE WITH THE PROTON TRANSFER INHIBITOR CD2+ | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CADMIUM ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2000-01-07 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the binding sites of the proton transfer inhibitors Cd2+ and Zn2+ in bacterial reaction centers.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5K47
 
 | | CryoEM structure of the human Polycystin-2/PKD2 TRP channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Pike, A.C.W, Grieben, M, Shintre, C.A, Tessitore, A, Shrestha, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2).
Nat. Struct. Mol. Biol., 24, 2017
|
|
1DY9
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1E3M
 
 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | Deposit date: | 2000-06-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
6JQD
 
 | | The structural basis of the beta-carbonic anhydrase CafC (L25G and L78G mutant) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
5NB7
 
 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NIW
 
 | | Glucose oxydase mutant A2 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hoffmann, K, Frank, D. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Shuffling Active Site Substate Populations Affects Catalytic Activity: The Case of Glucose Oxidase.
ACS Catal, 7, 2017
|
|
5NKS
 
 | | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4) | | Descriptor: | Dihydropyrimidinase-related protein 4 | | Authors: | Mathea, S, Elkins, J.M, Strain-Damerell, C, Salah, E, Borkowska, O, Chalk, R, Burgess-Brown, N, Pinkas, D.M, von Delft, F, Krojer, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2017-03-31 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4)
To Be Published
|
|
