3WOA
 
 | |
3SCG
 
 | | Fe(II)-HppE with R-HPP | | Descriptor: | Epoxidase, FE (II) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
3SCF
 
 | | Fe(II)-HppE with S-HPP and NO | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, Epoxidase, FE (II) ION, ... | | Authors: | Drennan, C.L. | | Deposit date: | 2011-06-07 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
J.Am.Chem.Soc., 133, 2011
|
|
4FSC
 
 | | Crystal Structure of Bacillus thuringiensis PlcR in its apo form | | Descriptor: | Transcriptional activator PlcR protein | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2012-06-27 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1Y9Q
 
 | |
3U3W
 
 | | Crystal Structure of Bacillus thuringiensis PlcR in complex with the peptide PapR7 and DNA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*AP*AP*AP*TP*AP*TP*TP*GP*CP*AP*TP*AP*G)-3', 5'-D(P*CP*TP*AP*TP*GP*CP*AP*AP*TP*AP*TP*TP*TP*CP*AP*TP*AP*T)-3', C-terminus heptapeptide from PapR protein, ... | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2011-10-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZKC
 
 | | Crystal structure of the master regulator for biofilm formation SinR in complex with DNA. | | Descriptor: | 5'-D(*AP*AP*AP*GP*TP*TP*CP*TP*CP*TP*TP*TP*AP*GP *AP*GP*AP*AP*CP*AP*AP)-3', 5'-D(*AP*TP*TP*GP*TP*TP*CP*TP*CP*TP*AP*AP*AP*GP *AP*GP*AP*AP*CP*TP*TP)-3', HTH-TYPE TRANSCRIPTIONAL REGULATOR SINR | | Authors: | Newman, J.A, Rodrigues, C, Lewis, R.J. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Basis of the Activity of Sinr, the Master Regulator of Biofilm Formation in Bacillus Subtilis.
J.Biol.Chem., 288, 2013
|
|
5YCL
 
 | |
2WIU
 
 | | Mercury-modified bacterial persistence regulator hipBA | | Descriptor: | CHLORIDE ION, HTH-TYPE TRANSCRIPTIONAL REGULATOR HIPB, MERCURY (II) ION, ... | | Authors: | Evdokimov, A, Voznesensky, I, Fennell, K, Anderson, M, Smith, J.F, Fisher, D.A. | | Deposit date: | 2009-05-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New Kinase Regulation Mechanism Found in Hipba: A Bacterial Persistence Switch.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6AF4
 
 | |
6AF3
 
 | |
4PU3
 
 | | Shewanella oneidensis MR-1 Toxin Antitoxin System HipA, HipB and its operator DNA complex (space group P212121) | | Descriptor: | Operator DNA, Toxin-antitoxin system antidote transcriptional repressor Xre family, Toxin-antitoxin system toxin HipA family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
4RYK
 
 | | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e | | Descriptor: | DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, Lmo0325 protein, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-15 | | Release date: | 2015-01-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e
To be Published
|
|
4PU4
 
 | | Shewanella oneidensis MR-1 Toxin Antitoxin System HipA, HipB and its operator DNA complex (space group P21) | | Descriptor: | Operator DNA, Toxin-antitoxin system antidote transcriptional repressor Xre family, Toxin-antitoxin system toxin HipA family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.786 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
3HZI
 
 | | Structure of mdt protein | | Descriptor: | 5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3', ADENOSINE-5'-TRIPHOSPHATE, HTH-type transcriptional regulator hipB, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
2QFC
 
 | | Crystal Structure of Bacillus thuringiensis PlcR complexed with PapR | | Descriptor: | C-terminus pentapeptide from PapR protein, DI(HYDROXYETHYL)ETHER, PlcR protein | | Authors: | Declerck, N, Chaix, D, Rugani, N, Hoh, F, Arold, S.T. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2P5T
 
 | |
3IVP
 
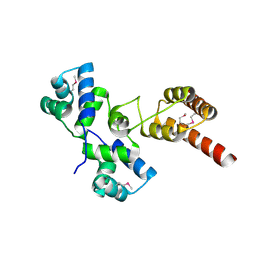 | |
2GRM
 
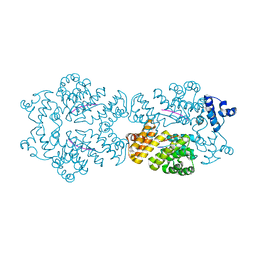 | | Crystal structure of PrgX/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
2GRL
 
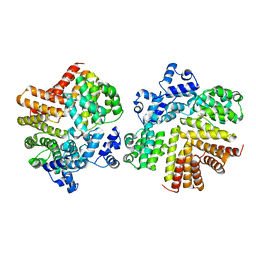 | | Crystal structure of dCT/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
1RIO
 
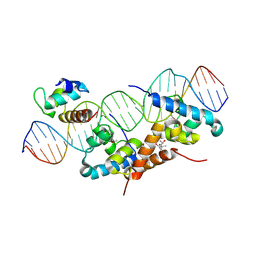 | | Structure of bacteriophage lambda cI-NTD in complex with sigma-region4 of Thermus aquaticus bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 27-MER, CALCIUM ION, ... | | Authors: | Jain, D, Nickels, B.E, Sun, L, Hochschild, A, Darst, S.A. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a ternary transcription activation complex.
Mol.Cell, 13, 2004
|
|
6HPC
 
 | |
6HPB
 
 | |
6KML
 
 | | 2.09 Angstrom resolution crystal structure of tetrameric HigBA toxin-antitoxin complex from E.coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
6KMQ
 
 | | 2.3 Angstrom resolution structure of dimeric HigBA toxin-antitoxin complex from E. coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
