3PMR
 
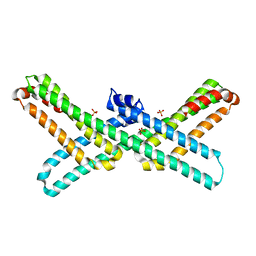 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
3PHQ
 
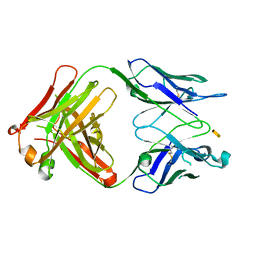 | | Crystal structure of S64-4 in complex with KDO | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, S64-4 Fab (IgG1) heavy chain, S64-4 Fab (IgG1) light chain | | Authors: | Evans, D.W, Evans, S.V. | | Deposit date: | 2010-11-04 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into parallel strategies for germline antibody recognition of lipopolysaccharide from Chlamydia.
Glycobiology, 21, 2011
|
|
3PND
 
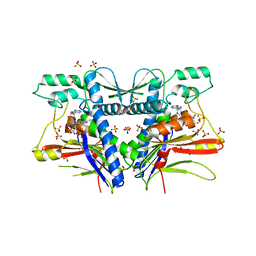 | | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD binding proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Boyd, J.M, Endrizzi, J.A, Hamilton, T.L, Christopherson, M.R, Mulder, D.W, Downs, D.M, Peters, J.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins.
J.Bacteriol., 193, 2011
|
|
3PJP
 
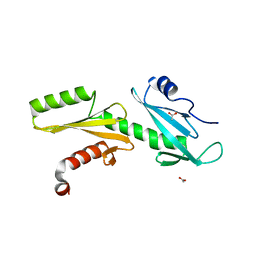 | | A Tandem SH2 Domain in Transcription Elongation Factor Spt6 Binds the Phosphorylated RNA Polymerase II C-terminal Repeat Domain(CTD) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Transcription elongation factor SPT6 | | Authors: | Sun, M, Lariviere, L, Dengl, S, Mayer, A, Cramer, P. | | Deposit date: | 2010-11-10 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD).
J.Biol.Chem., 285, 2010
|
|
3PJZ
 
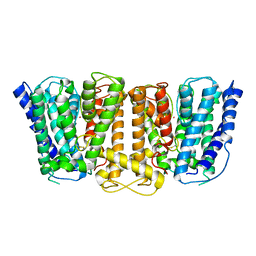 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
3Q2Y
 
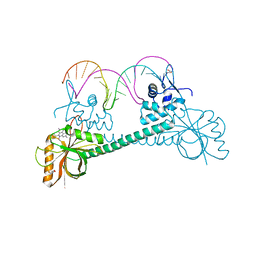 | | Crystal Structure of BmrR bound to ethidium | | Descriptor: | 23 bp promoter, ETHIDIUM, GLYCEROL, ... | | Authors: | Bachas, S, Eginton, C, Gunio, G, Wade, H. | | Deposit date: | 2010-12-20 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural contributions to multidrug recognition in the multidrug resistance (MDR) gene regulator, BmrR.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q3X
 
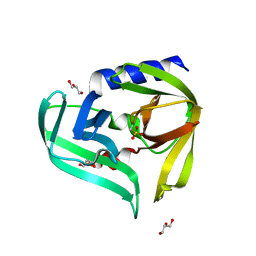 | | Crystal structure of the main protease (3C) from human enterovirus B EV93 | | Descriptor: | GLYCEROL, HEVB EV93 3C protease, MAGNESIUM ION | | Authors: | Costenaro, L, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3Q50
 
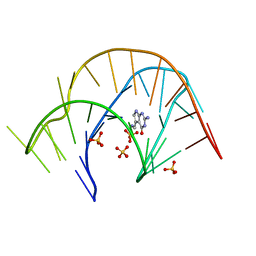 | |
3PKW
 
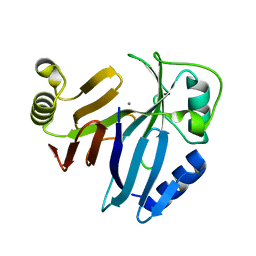 | | Crystal Structure of Toxoflavin Lyase (TflA) bound to Mn(II) | | Descriptor: | MANGANESE (II) ION, Toxoflavin lyase (TflA) | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toxoflavin lyase requires a novel 1-his-2-carboxylate facial triad .
Biochemistry, 50, 2011
|
|
3PLZ
 
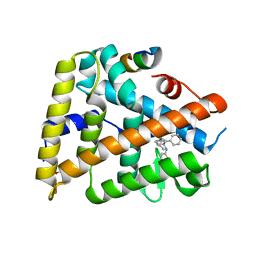 | | Human LRH1 LBD bound to GR470 | | Descriptor: | (3aS,6aR)-5-[(4E)-oct-4-en-4-yl]-N,4-diphenyl-2,3,6,6a-tetrahydropentalen-3a(1H)-amine, 1,2-ETHANEDIOL, FTZ-F1 related protein, ... | | Authors: | Williams, S.P, Xu, R, Zuercher, W.J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Agonists of the Orphan Nuclear Receptors Steroidogenic Factor-1 (SF-1, NR5A1) and Liver Receptor Homologue-1 (LRH-1, NR5A2).
J.Med.Chem., 54, 2011
|
|
3PMS
 
 | |
3PN1
 
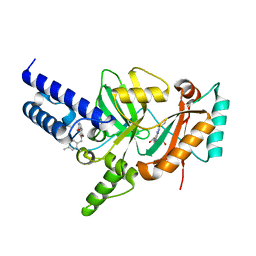 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | Authors: | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
3PNK
 
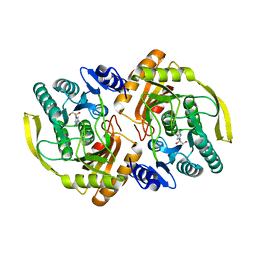 | | Crystal Structure of E.coli Dha kinase DhaK | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNQ
 
 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) complex with Dha | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PQ3
 
 | | Structure of I274C variant of E. coli KatE[] - Images 7-12 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PRJ
 
 | |
3PTZ
 
 | |
3PR3
 
 | | Crystal structure of Plasmodium falciparum glucose-6-phosphate isomerase (PF14_0341) in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Gileadi, T, Wernimont, A.K, Hutchinson, A, Weadge, J, Cossar, D, Lew, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Plasmodium falciparum glucose-6-phosphate isomerase (PF14_0341) in complex with fructose-6-phosphate
To be Published
|
|
3PSK
 
 | |
3PX1
 
 | | Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Strontium | | Descriptor: | Calcium-binding protein, STRONTIUM ION | | Authors: | Kumar, S, Kumar, S, Ahmad, E, Khan, R.H, Gourinath, S. | | Deposit date: | 2010-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Flexibility and plasticity of EF-hand motifs: Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Pb2+, Ba2+, and Sr2+.
To be Published
|
|
3PV2
 
 | |
3PXU
 
 | |
3PVT
 
 | |
3PW8
 
 | |
3PWQ
 
 | |
