6GXC
 
 | | Bacterial oligosaccharyltransferase PglB in complex with an inhibitory peptide and a reactive lipid-linked oligosaccharide analog | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLY-ASP-GLN-DAB-ALA-THR-PPN-GLY, ... | | Authors: | Napiorkowska, M, Locher, K.P, Boilevin, J, Darbre, T, Reymond, J.-L. | | Deposit date: | 2018-06-27 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structure of bacterial oligosaccharyltransferase PglB bound to a reactive LLO and an inhibitory peptide.
Sci Rep, 8, 2018
|
|
6GXK
 
 | | Crystal structure of Aldo-Keto Reductase 1C3 (AKR1C3) complexed with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 4-[[1-(4-chlorophenyl)carbonyl-5-methoxy-2-methyl-indol-3-yl]methyl]-1,2,5-oxadiazol-3-one, Aldo-keto reductase family 1 member C3, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bioisosteres of Indomethacin as Inhibitors of Aldo-Keto Reductase 1C3.
Acs Med.Chem.Lett., 10, 2019
|
|
6D68
 
 | | Ube2G1 in complex with ubiquitin variant Ubv.G1.1 | | Descriptor: | Ubiquitin-conjugating enzyme E2 G1, Ubv.G1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-20 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
7KNB
 
 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-09 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
5T3M
 
 | |
5T42
 
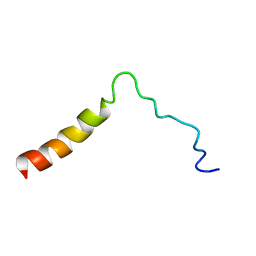 | | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity | | Descriptor: | Envelope glycoprotein | | Authors: | Lee, J, Nyenhuis, D.A, Nelson, E.A, Cafiso, D.S, White, J.M, Tamm, L.K. | | Deposit date: | 2016-08-28 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3BLU
 
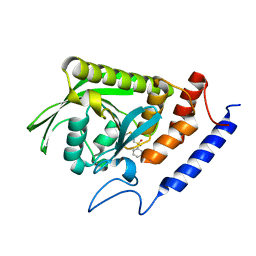 | | crystal structure YopH complexed with inhibitor PVS | | Descriptor: | (ethenylsulfonyl)benzene, Tyrosine-protein phosphatase yopH | | Authors: | Liu, S, Zhang, Z.-Y. | | Deposit date: | 2007-12-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aryl vinyl sulfonates and sulfones as active site-directed and mechanism-based probes for protein tyrosine phosphatases
J.Am.Chem.Soc., 130, 2008
|
|
5UYP
 
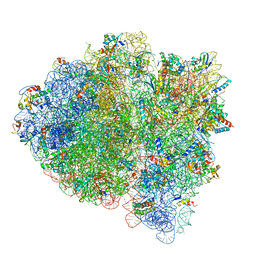 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, open 30S (Structure II-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
6DBQ
 
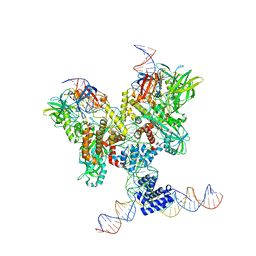 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DAM
 
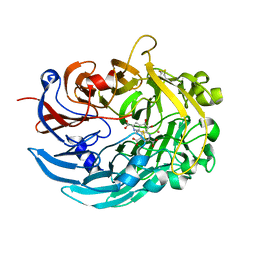 | | Crystal structure of lanthanide-dependent methanol dehydrogenase XoxF from Methylomicrobium buryatense 5G | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Deng, Y, Ro, S.Y, Rosenzweig, A.C. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C.
J. Biol. Inorg. Chem., 23, 2018
|
|
6H04
 
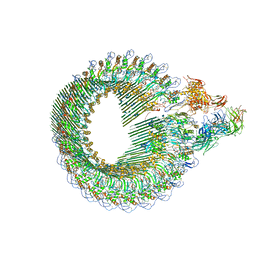 | | Closed conformation of the Membrane Attack Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
7KY7
 
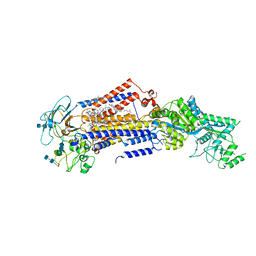 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the apo E1 state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
6DBW
 
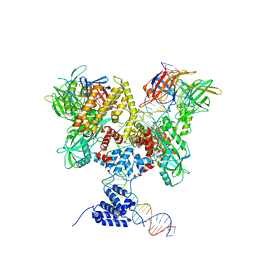 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5UNW
 
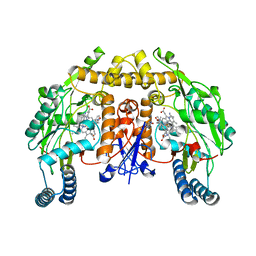 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 3-[2-(2-amino-4-methylquinolin-7-yl)ethyl]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 3-[2-(2-amino-4-methylquinolin-7-yl)ethyl]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UO8
 
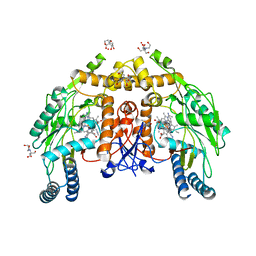 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 3-[(2-aminoquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-aminoquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
6H5A
 
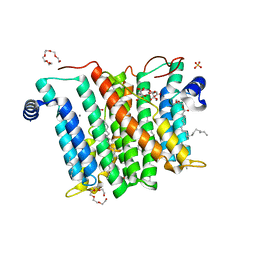 | |
6H5W
 
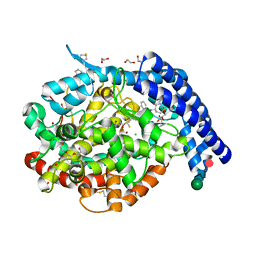 | |
7KGN
 
 | | S. Typhi YcbB - ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, L,D-transpeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2020-10-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Cellular Insights into the l,d-Transpeptidase YcbB as a Therapeutic Target in Citrobacter rodentium, Salmonella Typhimurium, and Salmonella Typhi Infections.
Antimicrob.Agents Chemother., 65, 2021
|
|
2P4Q
 
 | | Crystal Structure Analysis of Gnd1 in Saccharomyces cerevisiae | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating 1, CITRATE ANION | | Authors: | He, W, Wang, Y, Liu, W, Zhou, C.Z. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1
Bmc Struct.Biol., 7, 2007
|
|
7KJ3
 
 | | SARS-CoV-2 Spike Glycoprotein with two ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2P5R
 
 | | Crystal structure of the poplar glutathione peroxidase 5 in the oxidized form | | Descriptor: | CALCIUM ION, Glutathione peroxidase 5 | | Authors: | Koh, C.S, Didierjean, C, Navrot, N, Panjikar, S, Mulliert, G, Rouhier, N, Jacquot, J.-P, Aubry, A, Shawkataly, O, Corbier, C. | | Deposit date: | 2007-03-16 | | Release date: | 2007-07-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes.
J.Mol.Biol., 370, 2007
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
7KPS
 
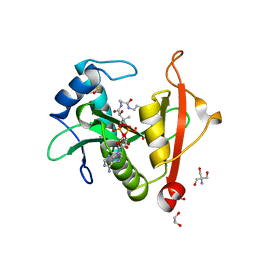 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
3AYC
 
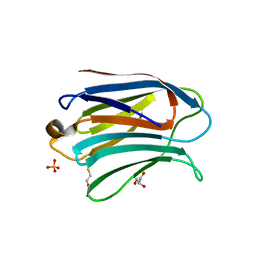 | | Crystal structure of galectin-3 CRD domian complexed with GM1 pentasaccharide | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Galectin-3, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
7KNH
 
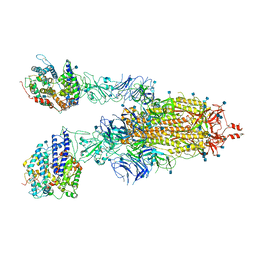 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
