6VBV
 
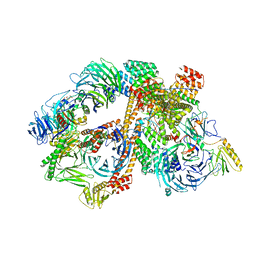 | | Structure of the bovine BBSome:ARL6:GTP complex | | Descriptor: | ADP-ribosylation factor-like protein 6, BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, ... | | Authors: | Singh, S.K, Gui, M, Koh, F, Yip, M.C.J, Brown, A. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and activation mechanism of the BBSome membrane protein trafficking complex.
Elife, 9, 2020
|
|
5EY7
 
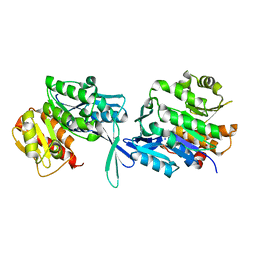 | |
6XKS
 
 | | Crystal structure of domain A from the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) of Salmonella typhimurium | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Histidine ABC transporter substrate-binding protein HisJ | | Authors: | Romero-Romero, S, Berrocal, T, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermodynamic and kinetic analysis of the LAO binding protein and its isolated domains reveal non-additivity in stability, folding and function.
Febs J., 2023
|
|
6XQV
 
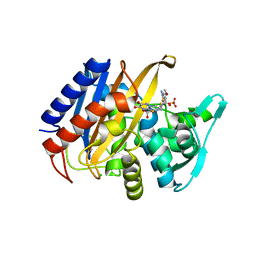 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | Descriptor: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6VBW
 
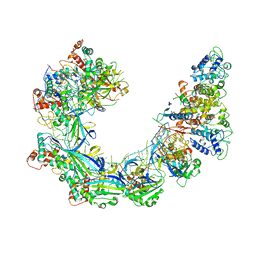 | |
6XQZ
 
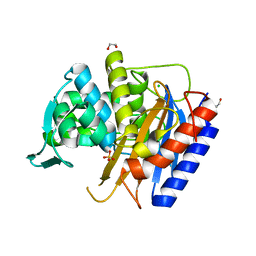 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
4OD5
 
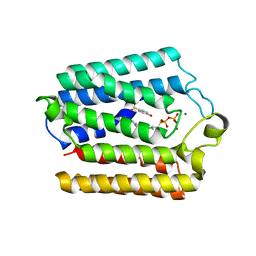 | |
6XQY
 
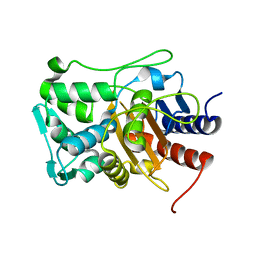 | |
6XQX
 
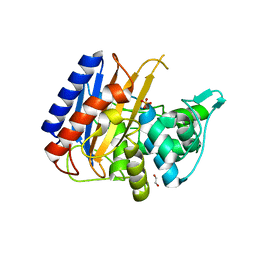 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
1XPG
 
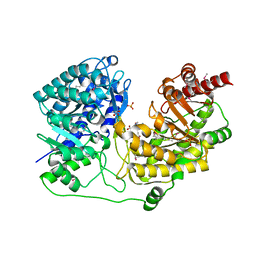 | | Crystal Structure of T. maritima Cobalamin-Independent Methionine Synthase complexed with Zn2+ and Methyltetrahydrofolate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, MESO-ERYTHRITOL, ... | | Authors: | Pejchal, R, Ludwig, M.L. | | Deposit date: | 2004-10-08 | | Release date: | 2005-03-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication
Plos Biol., 3, 2005
|
|
6XTF
 
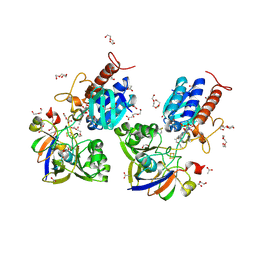 | | Crystal structure a Thioredoxin Reductase from Gloeobacter violaceus bound to its electron donor | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Buey, R.M, Gonzalez-Holgado, G, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria.
Plant Physiol., 186, 2021
|
|
6WUI
 
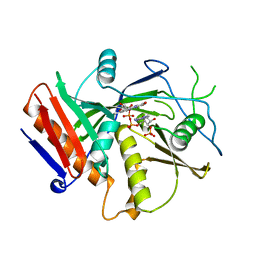 | | Crystal Structure of mutant S. pombe Rai1 (E150S/E199Q/E239Q) in complex with 3'-FADP | | Descriptor: | Decapping nuclease din1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs.
Nucleic Acids Res., 48, 2020
|
|
6WXW
 
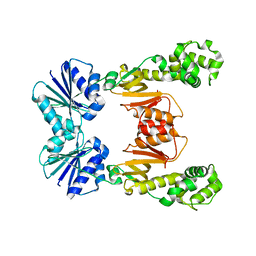 | |
5GHJ
 
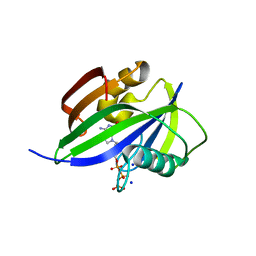 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHP
 
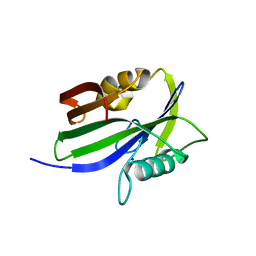 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.192 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
6XYZ
 
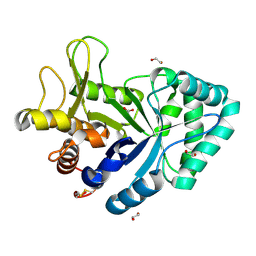 | | Crystal structure of the GH18 chitinase ChiB from the chitin utilization locus of Flavobacterium johnsoniae | | Descriptor: | 1,2-ETHANEDIOL, Candidate chitinase Glycoside hydrolase family 18, FORMIC ACID | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V, Pope, P, Branden, G, Larsbrink, J. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
6XB3
 
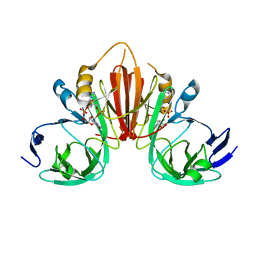 | |
1DUP
 
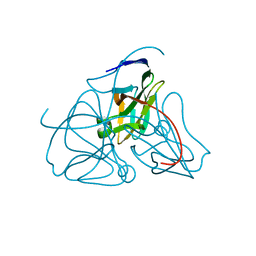 | | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE (D-UTPASE) | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Wilson, K.S, Larsson, G, Nyman, P.O, Cedergren, E. | | Deposit date: | 1995-09-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dUTPase.
Nature, 355, 1992
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
5ENK
 
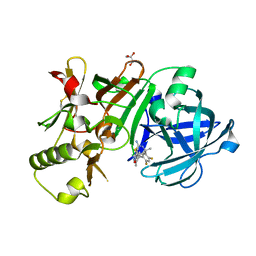 | | Compound 18 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)-5-pyrimidin-5-yl-phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Lewis, H.A. | | Deposit date: | 2015-11-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Targeting the BACE1 Active Site Flap Leads to a Potent Inhibitor That Elicits Robust Brain A beta Reduction in Rodents.
Acs Med.Chem.Lett., 7, 2016
|
|
4DVF
 
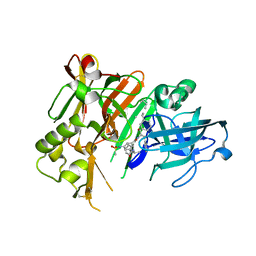 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
5TGA
 
 | |
5ETI
 
 | | Structure of dead kinase MAPK14 | | Descriptor: | Mitogen-activated protein kinase 14 | | Authors: | Pellegrini, E, Bowler, M.W. | | Deposit date: | 2015-11-17 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the MKK6-p38 alpha complex defines the basis of MAPK specificity and activation.
Science, 381, 2023
|
|
5ETC
 
 | |
6XG8
 
 | | ISCth4 transposase, pre-cleaved complex, PCC | | Descriptor: | DNA (26-MER), Mutator family transposase | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
