6LQB
 
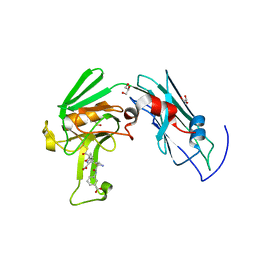 | | A third intermediate state of L,D-transpeptidase LdtMt2-ertapenem adduct | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, D.F, Qin, Y.L. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New State I-plus Observed for the L,D-transpeptidase LdtMt2-ertapenem Adduct
Prog.Biochem.Biophys., 47, 2020
|
|
6LQH
 
 | | High resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6LN7
 
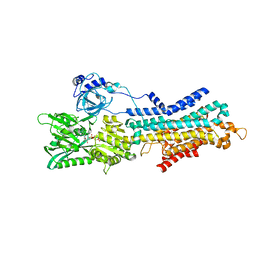 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class3) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LNK
 
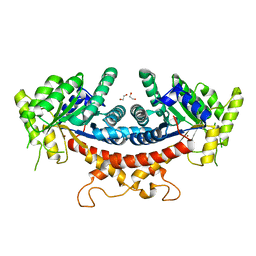 | | Candida albicans Fructose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Huang, Y, Cao, H, Ren, Y, Wan, J. | | Deposit date: | 2019-12-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
6LNW
 
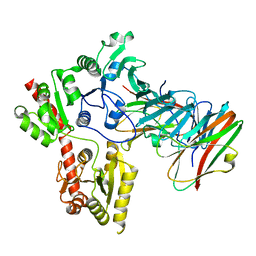 | | Crystal structure of accessory secretory protein 1,2 and 3 in Streptococcus pneumoniae | | Descriptor: | Accessory secretory protein Asp1, Accessory secretory protein Asp2, Accessory secretory protein Asp3 | | Authors: | Guo, C, Feng, Z, Zuo, G. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the Asp1/2/3 complex mediated secretion of pneumococcal serine-rich repeat protein PsrP.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6LRG
 
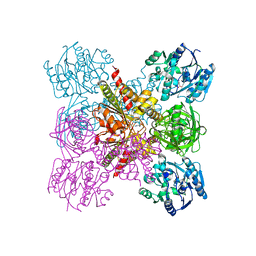 | | Crystal Structure of the Ternary Complex of AgrE with Ornithine and NAD+ | | Descriptor: | Alr4995 protein, L-ornithine, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, H, Rhee, S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41218114 Å) | | Cite: | Structural and mutational analyses of the bifunctional arginine dihydrolase and ornithine cyclodeaminase AgrE from the cyanobacteriumAnabaena.
J.Biol.Chem., 295, 2020
|
|
6LRR
 
 | | Cryo-EM structure of RuBisCO-Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6LSZ
 
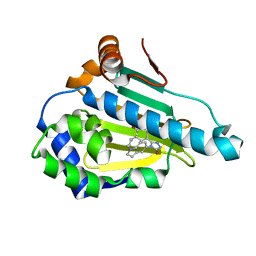 | | HSP 90 in complex with Ganetespib | | Descriptor: | 5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(1-methyl-1H-indol-5-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure of HSP90 in complex with Ganetespib
To Be Published
|
|
6LT4
 
 | | AAA+ ATPase, ClpL from Streptococcus pneumoniae: ATPrS-bound | | Descriptor: | ATP-dependent Clp protease, ATP-binding subunit, MAGNESIUM ION, ... | | Authors: | Kim, G, Lee, S.G, Han, S, Jung, J, Jeong, H.S, Hyun, J.K, Rhee, D.K, Kim, H.M, Lee, S. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Faseb J., 34, 2020
|
|
6LTU
 
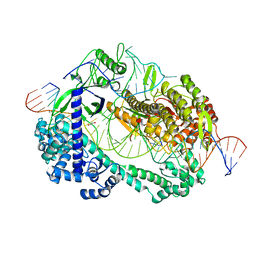 | | Crystal structure of Cas12i2 ternary complex with double Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LU2
 
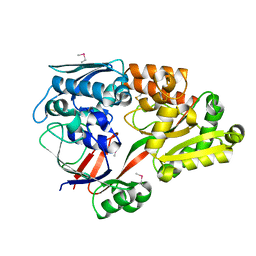 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans | | Descriptor: | Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LUB
 
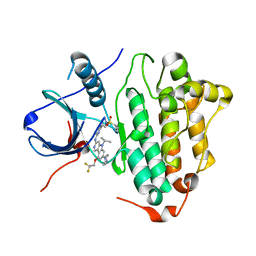 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with CH7233163 | | Descriptor: | Epidermal growth factor receptor, N-[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]-7-(4-methylpiperazin-1-yl)-5-propan-2-yl-9-[2,2,2-tris(fluoranyl)ethoxy]pyrido[4,3-b]indol-3-amine | | Authors: | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | Deposit date: | 2020-01-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
6LUH
 
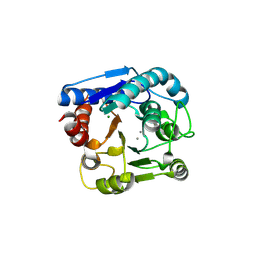 | | High resolution structure of N(omega)-hydroxy-L-arginine hydrolase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, N(omega)-hydroxy-L-arginine amidinohydrolase | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-09-02 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an Nomega-hydroxy-L-arginine hydrolase found in the D-cycloserine biosynthetic pathway.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6LOM
 
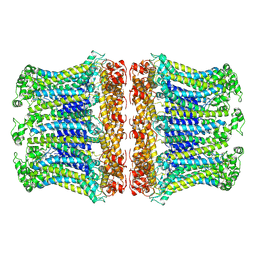 | |
6LQ8
 
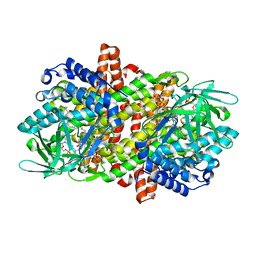 | |
6LUM
 
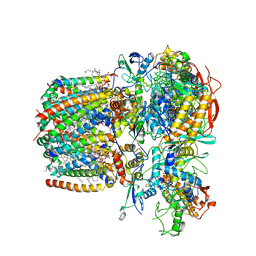 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
6LV9
 
 | | Cu- Carbonic Anhydrase II pH 7.8 0 atm CO2 | | Descriptor: | COPPER (II) ION, Carbonic anhydrase 2, GLYCEROL | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2020-02-02 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Elucidating the role of metal ions in carbonic anhydrase catalysis.
Nat Commun, 11, 2020
|
|
6LVL
 
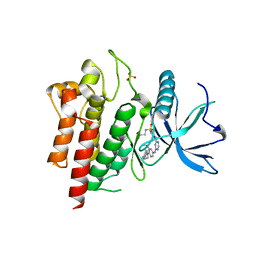 | | Crystal structure of FGFR2 in complex with 1,3,5-triazine derivative | | Descriptor: | Fibroblast growth factor receptor 2, N-ethyl-2-[[4-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-1,3,5-triazin-2-yl]amino]benzenesulfonamide, SULFATE ION | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
6LVN
 
 | | Structure of the 2019-nCoV HR2 Domain | | Descriptor: | Spike protein S2' | | Authors: | Zhu, Y, Sun, F. | | Deposit date: | 2020-02-04 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of HR2 domain of 2019-nCoV S2 subunit
To Be Published
|
|
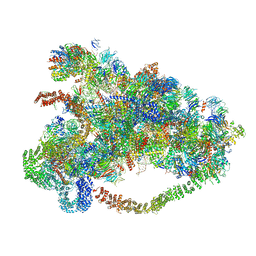
6LQR
 
 | |
6LVW
 
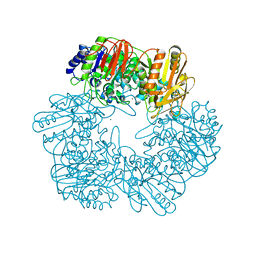 | |
6LX0
 
 | |
6LX4
 
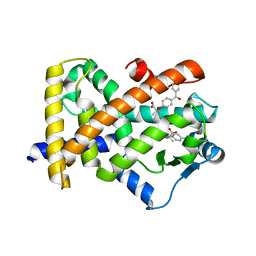 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXD
 
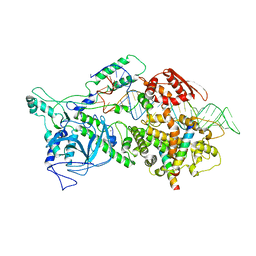 | | Pri-miRNA bound DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, RNA (102-mer), Ribonuclease 3, ... | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
6LW5
 
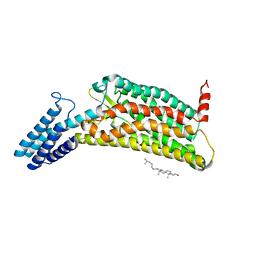 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
