1DLH
 
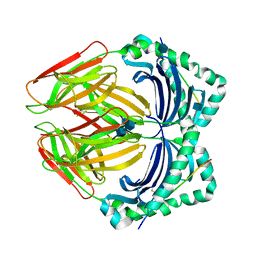 | |
6ZO8
 
 | | Minocycline binding to the deep binding pocket of AcrB-G621P | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
8VK6
 
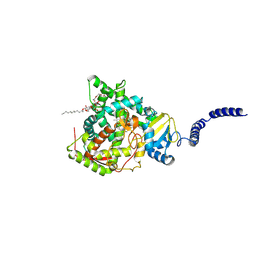 | | 14alpha-demethylase (CYP51) with amide-linked long arm extension antifungal azole inhibitor | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Lanosterol 14-alpha demethylase, N-[(2S)-2-(2,4-difluorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]-4'-(trifluoromethoxy)[1,1'-biphenyl]-4-carboxamide, ... | | Authors: | Tyndall, J.D.A, Monk, B.C, Simons, C. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Exploring Long Arm Amide-Linked Side Chains in the Design of Antifungal Azole Inhibitors of Sterol 14 alpha-Demethylase (CYP51).
J.Med.Chem., 68, 2025
|
|
4MZC
 
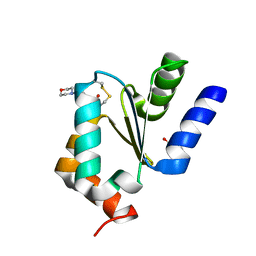 | | Atomic Resolution Structure of PfGrx1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Glutaredoxin | | Authors: | Yogavel, M, Sharma, A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (0.949 Å) | | Cite: | Atomic resolution crystal structure of glutaredoxin 1 from Plasmodium falciparum and comparison with other glutaredoxins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8VMI
 
 | | PRC2_AJ119-450 bound to H3K4me3 | | Descriptor: | EZH2, Histone H3.1, Histone H3.1t, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6ZO7
 
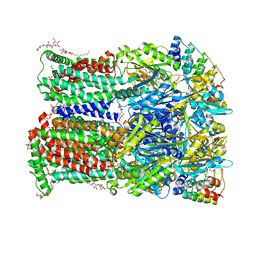 | | 3-Formylrifamycin SV binding to the access pocket of AcrB-G619P L and T protomer | | Descriptor: | (2S,12Z,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-8-formyl-5,6,9,17,19-pentahydroxy-23-methoxy-2,4,12,16,18,20,22-heptam ethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-21-yl acetate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
6ZOH
 
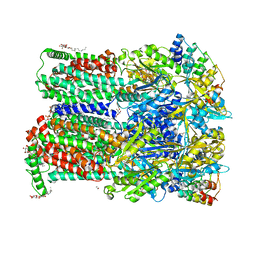 | | 3-Formylrifamycin SV binding to the access pocket of AcrB-G619P_G621P L and T protomers | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2S,12Z,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-8-formyl-5,6,9,17,19-pentahydroxy-23-methoxy-2,4,12,16,18,20,22-heptam ethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-21-yl acetate, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
8VVE
 
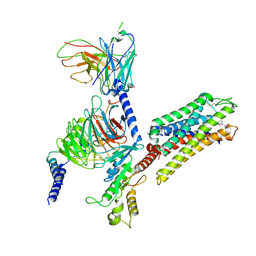 | | Kappa opioid receptor:Galphai protein in complex with inverse agonist norBNI | | Descriptor: | (4bS,8R,8aS,10aS,11R,14aS,19aR,20bR)-7,12-bis(cyclopropylmethyl)-5,6,7,8,9,10,11,12,13,14,20,20b-dodecahydro-19aH-4,8:11,15-dimethanobis[1]benzofuro[2,3-a:3',2'-i]dipyrido[4,3-b:3',4'-h]carbazole-1,8a,10a,18-tetrol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gati, C, Motiwala, Z, Tyson, A.S, Styrpejko, D, Han, G.W, Khan, S, Ramos-Gonzalez, N, Shenvi, R, Majumdar, S. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanisms of inverse agonism via kappa-opioid receptor-G protein complexes.
Nat.Chem.Biol., 21, 2025
|
|
7A5Y
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with Rp-dGTP-alphaS (T8T) and Mg | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Purkiss, A, Taylor, I.A. | | Deposit date: | 2020-08-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Probing the Catalytic Mechanism and Inhibition of SAMHD1 Using the Differential Properties of R p - and S p -dNTP alpha S Diastereomers.
Biochemistry, 60, 2021
|
|
1DFV
 
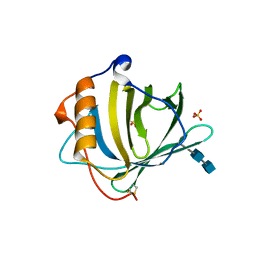 | | CRYSTAL STRUCTURE OF HUMAN NEUTROPHIL GELATINASE ASSOCIATED LIPOCALIN MONOMER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN NEUTROPHIL GELATINASE, ... | | Authors: | Goetz, D.H, Willie, S.T, Armen, R.S, Bratt, T, Borregaard, N, Strong, R.K. | | Deposit date: | 1999-11-22 | | Release date: | 2000-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin
Biochemistry, 39, 2000
|
|
3GVL
 
 | | Crystal Structure of endo-neuraminidaseNF | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
1DDX
 
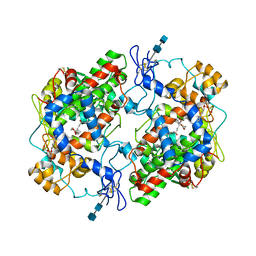 | | CRYSTAL STRUCTURE OF A MIXTURE OF ARACHIDONIC ACID AND PROSTAGLANDIN BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2: PROSTAGLANDIN STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[6-(3-HYDROPEROXY-OCT-1-ENYL)-2,3-DIOXA-BICYCLO[2.2.1]HEPT-5-YL]-HEPT-5-ENOIC ACID, ... | | Authors: | Kiefer, J.R, Pawlitz, J.L, Moreland, K.T, Stegeman, R.A, Gierse, J.K, Stevens, A.M, Goodwin, D.C, Rowlinson, S.W, Marnett, L.J, Stallings, W.C, Kurumbail, R.G. | | Deposit date: | 1999-11-11 | | Release date: | 2000-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the stereochemistry of the cyclooxygenase reaction.
Nature, 405, 2000
|
|
8VML
 
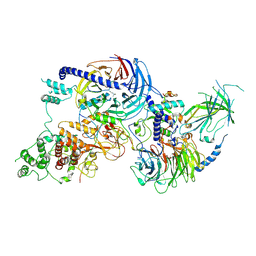 | | PRC2_AJ1-450 bound to H3K4me3 | | Descriptor: | AEPB2, EED, EZH2, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VNV
 
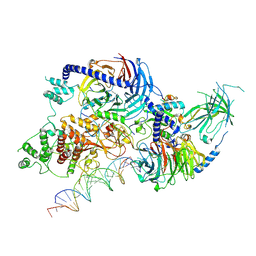 | |
3GXN
 
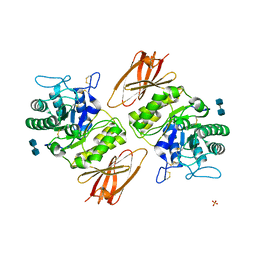 | | Crystal structure of apo alpha-galactosidase A at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, SULFATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
4MW0
 
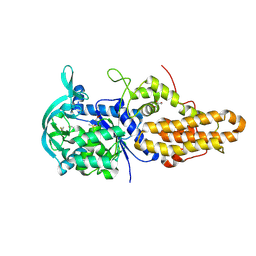 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea (Chem 1392) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
8VNZ
 
 | |
8VVF
 
 | | Kappa opioid receptor:Galphai protein in complex with inverse agonist JDTic | | Descriptor: | (3R)-7-hydroxy-N-{(2S)-1-[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]-3-methylbutan-2-yl}-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gati, C, Motiwala, Z, Tyson, A.S, Styrpejko, D, Han, G.W, Khan, S, Ramos-Gonzalez, N, Shenvi, R, Majumdar, S. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanisms of inverse agonism via kappa-opioid receptor-G protein complexes.
Nat.Chem.Biol., 21, 2025
|
|
8VVG
 
 | | Kappa opioid receptor in complex with heterotrimerig Gi protein, bound to inverse agonist GB18 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Gati, C, Motiwala, Z, Tyson, A.S, Styrpejko, D, Han, G.W, Khan, S, Ramos-Gonzalez, N, Shenvi, R, Majumdar, S. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-15 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanisms of inverse agonism via kappa-opioid receptor-G protein complexes.
Nat.Chem.Biol., 21, 2025
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
7A53
 
 | | Structure of DYRK1A in complex with compound 7 | | Descriptor: | (3~{E})-3-[(5-methylfuran-2-yl)methylidene]-1~{H}-indol-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A4R
 
 | | Structure of DYRK1A in complex with compound 1 | | Descriptor: | 2-amino-3H,4H,7H-pyrrolo[2,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A4O
 
 | | Structure of DYRK1A in complex with AMPNP | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A55
 
 | | Structure of DYRK1A in complex with compound 8 | | Descriptor: | 3-(1-methylpyrazol-4-yl)-1~{H}-pyrazole-5-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A4S
 
 | | Structure of DYRK1A in complex with compound 2 | | Descriptor: | 7-chlorothieno[3,2-c]pyridin-4-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
