7O1G
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, Putative acyltransferase Rv0859, SULFATE ION | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1K
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4V
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with oxidized nicotinamide adenine dinucleotide | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
4YMK
 
 | | Crystal Structure of Stearoyl-Coenzyme A Desaturase 1 | | Descriptor: | Acyl-CoA desaturase 1, STEAROYL-COENZYME A, ZINC ION, ... | | Authors: | Bai, Y, McCoy, J.G, Rajashankar, K.R, Zhou, M. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | X-ray structure of a mammalian stearoyl-CoA desaturase.
Nature, 524, 2015
|
|
7BWR
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
8QC4
 
 | | M. tuberculosis salicylate synthase MbtI in complex with 5-(3-carboxyphenyl)furan-2-carboxylic acid | | Descriptor: | 5-(3-carboxyphenyl)furan-2-carboxylic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Mori, M, Villa, S, Meneghetti, F, Bellinzoni, M. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Structural Study of a New MbtI-Inhibitor Complex: Towards an Optimized Model for Structure-Based Drug Discovery.
Pharmaceuticals, 16, 2023
|
|
8R1F
 
 | | Monomeric E6AP-E6-p53 ternary complex | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakraborty, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
8R1G
 
 | | Dimeric ternary structure of E6AP-E6-p53 | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakrabory, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
4YTN
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[3-(pentafluorophenoxy)phenyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New Insights into the Design of Inhibitors Targeted for Parasitic Anaerobic Energy Metabolism
To Be Published
|
|
8QF2
 
 | | Beta-L-Arabinofurano-cyclitol Aziridines are Cysteine-directed Broad-spectrum Inhibitors and Activity-based Probes for Retaining Beta-L-arabinofuranosidases | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Borlandelli, V, Offen, W.A, Moroz, O, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
4YOP
 
 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Nango, E. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native sulfur/chlorine SAD phasing for serial femtosecond crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
8QND
 
 | | Crystal structure of the ribonucleoside hydrolase C from Lactobacillus reuteri | | Descriptor: | CALCIUM ION, Inosine-uridine nucleoside N-ribohydrolase | | Authors: | Matyuta, I.O, Minyaev, M.E, Shaposhnikov, L.A, Pometun, E.V, Tishkov, V.I, Popov, V.O, Boyko, K.M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Functional Examination of Novel Ribonucleoside Hydrolase C (RihC) from Limosilactobacillus reuteri LR1.
Int J Mol Sci, 25, 2023
|
|
4Y6V
 
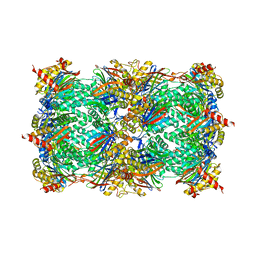 | | Yeast 20S proteasome in complex with Ac-PAE-ep | | Descriptor: | Ac-PAE-ep, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y84
 
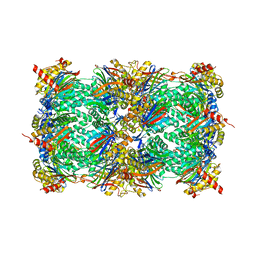 | | Yeast 20S proteasome in complex with N3-A(4,4-F2P)nLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y8U
 
 | |
7NRZ
 
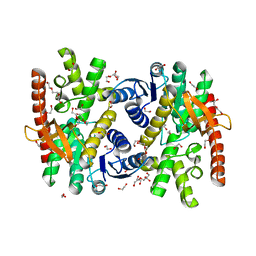 | | Crystal structure of malate dehydrogenase from Trypanosoma cruzi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sonani, R.R, Kurpiewska, K, Lewinski, K, Dubin, G. | | Deposit date: | 2021-03-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct sequence and structural feature of trypanosoma malate dehydrogenase.
Biochem.Biophys.Res.Commun., 557, 2021
|
|
4YAF
 
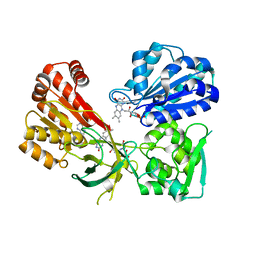 | | rat CYPOR with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4AL2
 
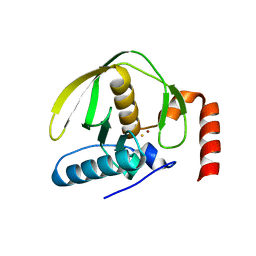 | |
7NVP
 
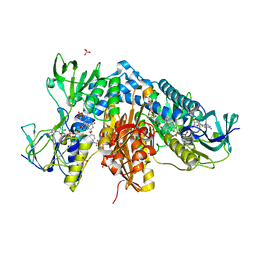 | | Trypanothione reductase from Trypanosoma brucei in complex with N-{4-methoxy-3-[(4-methoxyphenyl)sulfamoyl]phenyl}-5-nitrothiophene-2-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N(1),N(8)-bis(glutathionyl)spermidine reductase, ... | | Authors: | Battista, T, Fiorillo, A, Colotti, G, Ilari, A. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Optimization of Potent and Specific Trypanothione Reductase Inhibitors: A Structure-Based Drug Discovery Approach.
Acs Infect Dis., 8, 2022
|
|
4Y74
 
 | | Yeast 20S proteasome in complex with Ac-LAL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAL-ep, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
7NWV
 
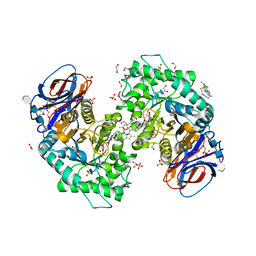 | | Structure of recombinant human beta-glucocerebrosidase in complex with BODIPY Tagged Cyclophellitol activity based probe | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},5~{S})-4-[[4-[4-[2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-3-aza-1-azonia-2-boranuidatricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaen-8-yl]butyl]-1,2,3-triazol-1-yl]methyl]cyclohexane-1,2,3,5-tetrol, 1,2-ETHANEDIOL, 1-deoxy-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
8SBI
 
 | |
4Y8M
 
 | | Yeast 20S proteasome beta7-delta7_Cter mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
3KLX
 
 | |
