3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OMG
 
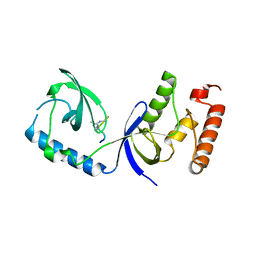 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R14me2s | | Descriptor: | Staphylococcal nuclease domain-containing protein 1, dimethylated arginine peptide R14me2s | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OV9
 
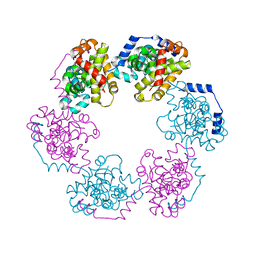 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein, SODIUM ION | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
3OVU
 
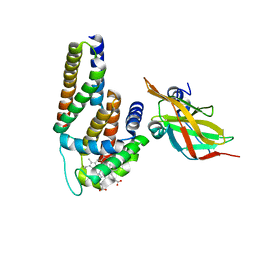 | | Crystal Structure of Human Alpha-Haemoglobin Complexed with AHSP and the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Alpha-hemoglobin-stabilizing protein, Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, ... | | Authors: | Jacques, D.A, Krishna Kumar, K, Caradoc-Davies, T.T, Langley, D.B, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A new haem pocket structure in alpha-haemoglobin
To be Published
|
|
3OWB
 
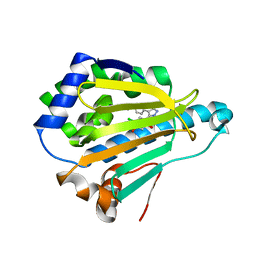 | | Crystal Structure of HSP90 with VER-49009 | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3ONM
 
 | | Effector binding Domain of LysR-Type transcription factor RovM from Y. pseudotuberculosis | | Descriptor: | Transcriptional regulator LrhA | | Authors: | Quade, N, Diekmann, M, Haffke, M, Heroven, A.K, Dersch, P, Heinz, D.W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the effector-binding domain of the LysR-type transcription factor RovM from Yersinia pseudotuberculosis.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OWU
 
 | |
3OX3
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(2-methoxy-1H-dipyrido[2,3-a:3',2'-e]pyrrolizin-11-yl)ethyl]furan-2-carboxamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3OO8
 
 | |
3OYK
 
 | | Crystal structure of the PFV S217H mutant intasome bound to manganese | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OZI
 
 | | Crystal structure of the TIR domain from the flax disease resistance protein L6 | | Descriptor: | COBALT (II) ION, L6tr | | Authors: | Ve, T, Bernoux, M, Williams, S, Valkov, E, Warren, C, Hatters, D, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2010-09-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation.
Cell Host Microbe, 9, 2011
|
|
3OQK
 
 | | Crystal Structure Analysis of Renin-indole-piperazin inhibitor complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-phenoxy-1-phenyl-3-(piperazin-1-ylcarbonyl)-1H-indole, GLYCEROL, ... | | Authors: | Bocskei, Z. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and optimization of a new class of potent and non-chiral indole-3-carboxamide-based renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3P10
 
 | |
3P2E
 
 | | Structure of an antibiotic related Methyltransferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 2010
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | Descriptor: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P3T
 
 | |
3P45
 
 | | Crystal structure of apo-caspase-6 at physiological pH | | Descriptor: | caspase-6 | | Authors: | Mueller, I, Lamers, M.B.A.C, Ritchie, A.J, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2010-10-06 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures of active and inhibitor-bound human Casp6
To be Published
|
|
3P5A
 
 | |
3P62
 
 | |
3P6M
 
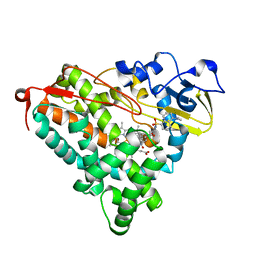 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans | | Descriptor: | ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans
To be Published
|
|
3OX9
 
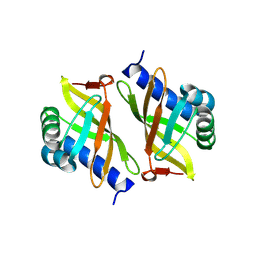 | |
3PA4
 
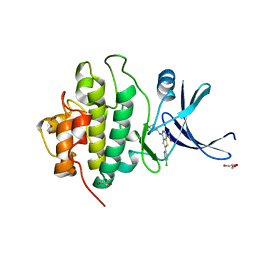 | | X-ray crystal structure of compound 2a bound to human CHK1 kinase domain | | Descriptor: | 2-(4-chlorophenyl)-4-[(3S)-piperidin-3-ylamino]thieno[3,2-c]pyridine-7-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OY4
 
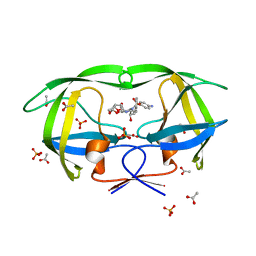 | | Crystal Structure of HIV-1 L76V Protease in Complex with the Protease Inhibitor Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 PROTEASE, ... | | Authors: | Schiffer, C.A, Nalivaika, E.A, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of HIV-1 L76V Protease in Complex with the Protease Inhibitor Darunavir.
To be Published
|
|
3OYC
 
 | |
3OYN
 
 | | Crystal structure of the PFV N224H mutant intasome bound to magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
