6Y16
 
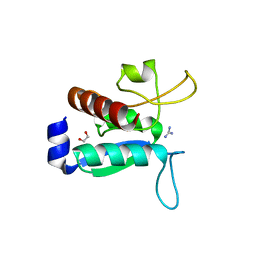 | | CRYSTAL STRUCTURE OF TMARGBP DOMAIN 1 IN COMPLEX WITH THE GUANIDINIUM ION | | Descriptor: | 1,2-ETHANEDIOL, Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, ... | | Authors: | Ruggiero, A, Balasco, N, Smaldone, G, Graziano, G, Vitagliano, L. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium binding to proteins: The intriguing effects on the D1 and D2 domains of Thermotoga maritima Arginine Binding Protein and a comprehensive analysis of the Protein Data Bank.
Int.J.Biol.Macromol., 163, 2020
|
|
6BBR
 
 | |
7PID
 
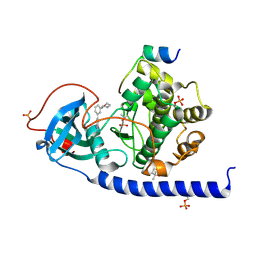 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN060 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(3R)-morpholin-3-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
6BC2
 
 | |
7PIE
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN068 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(2R)-morpholin-2-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.427 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
6BC7
 
 | | Cryo X-ray structure of sisomicin bound AAC-VIa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, AAC 3-VI protein, ACETATE ION | | Authors: | Cuneo, M.J, Kumar, P. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A low-barrier hydrogen bond mediates antibiotic resistance in a noncanonical catalytic triad.
Sci Adv, 4, 2018
|
|
7PIF
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN086 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[3-[4-(aminomethyl)oxan-4-yl]phenyl]-2-azanyl-benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIH
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN093 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[3-[[(3~{R})-3-azanylpyrrolidin-1-yl]methyl]phenyl]-4~{H}-isoquinolin-1-one, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIG
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN088 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-azanyl-2-chloranyl-3-[(Z)-piperidin-3-ylidenemethyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
6M6D
 
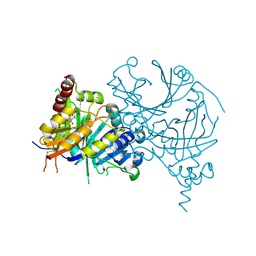 | | Structure of HPPD complexed with a synthesized inhibitor | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(3-trimethylsilylprop-2-ynyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Yang, W.-C, Yang, G.-F. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Structure-Guided Discovery of Silicon-Containing Subnanomolar Inhibitor of Hydroxyphenylpyruvate Dioxygenase as a Potential Herbicide.
J.Agric.Food Chem., 69, 2021
|
|
7PNS
 
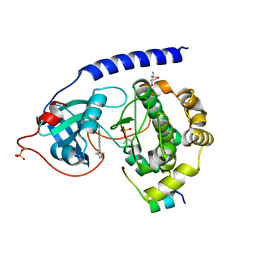 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN081 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-6-[5-[(dimethylamino)methyl]-2-fluoranyl-phenyl]-1H-indole-3-carbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
3IBJ
 
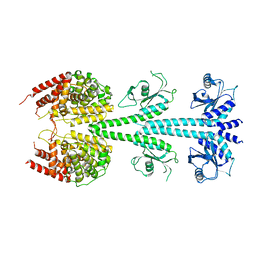 | | X-ray structure of PDE2A | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | Authors: | Pandit, J. | | Deposit date: | 2009-07-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6B44
 
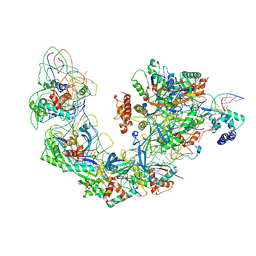 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound target dsDNA | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6M6T
 
 | | Amylomaltase from Streptococcus agalactiae in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Wangkanont, K, Tumhom, S, Pongsawasdi, P. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Streptococcus agalactiae amylomaltase offers insight into the transglycosylation mechanism and the molecular basis of thermostability among amylomaltases.
Sci Rep, 11, 2021
|
|
1IIY
 
 | |
6YND
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 1 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.525 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
8D9T
 
 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes, centered on gamma-Arf1 | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
6AP8
 
 | |
8D9S
 
 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated wide membrane tubes, centered on beta-Arf1 | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D9U
 
 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes, centered on beta-Arf1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D9R
 
 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated wide membrane tubes, centered on gamma-Arf1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
3IJX
 
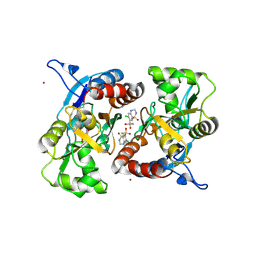 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydrochlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
6VC1
 
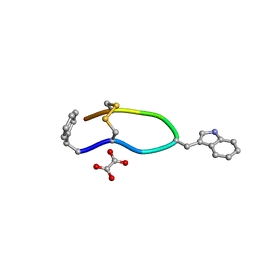 | | Octreotide oxalate | | Descriptor: | OXALATE ION, Octreotide | | Authors: | Spiliopoulou, M, Karavassili, F, Triandafillidis, D, Valmas, A, Kosinas, C, Fili, S, Barlos, K, Barlos, K.K, Morin, M, Reinle-Schmitt, M, Gozzo, F, Margiolaki, I. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2021-05-19 | | Method: | POWDER DIFFRACTION | | Cite: | New perspectives in macromolecular powder diffraction using single-photon-counting strip detectors: high-resolution structure of the pharmaceutical peptide octreotide.
Acta Crystallogr.,Sect.A, 77, 2021
|
|
3ITU
 
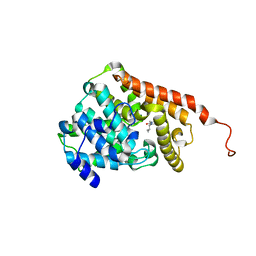 | | hPDE2A catalytic domain complexed with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4Q86
 
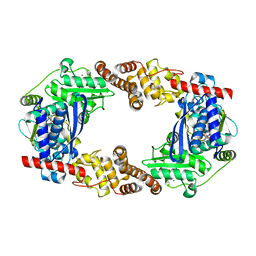 | | YcaO with AMP Bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
