4QIP
 
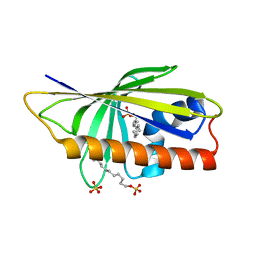 | |
5B1N
 
 | | DHp domain structure of EnvZ from Escherichia coli | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
4QI0
 
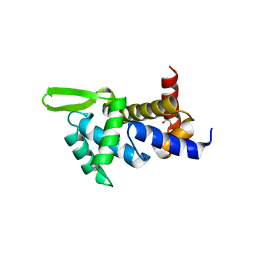 | | X-ray structure of the ROQ domain from murine Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4BMJ
 
 | | Structure of the UBZ1and2 tandem of the ubiquitin-binding adaptor protein TAX1BP1 | | Descriptor: | CHLORIDE ION, TAX1-BINDING PROTEIN 1, ZINC ION | | Authors: | Ceregido, M.A, Spinola-Amilibia, M, Buts, L, Rivera, J, Bravo, J, van Nuland, N.A.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of Tax1BP1 Ubz1 + 2 Provides Insight Into Target Specificity and Adaptability
J.Mol.Biol., 426, 2014
|
|
3Q7Z
 
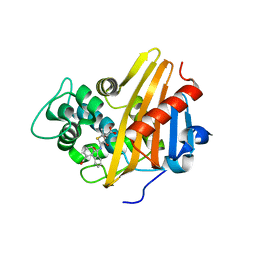 | | CBAP-acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2'-carboxybiphenyl-2-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase regulatory protein BlaR1 | | Authors: | Borbulevych, O.Y, Mobashery, S, Baker, B.M. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Lysine Nzeta-decarboxylation switch and activation of the beta-lactam sensor domain of BlaR1 protein of methicillin-resistant Staphylococcus aureus.
J.Biol.Chem., 286, 2011
|
|
3QBC
 
 | |
4QVX
 
 | |
4R0O
 
 | | Crystal structure of PEGylated plastocyanin at 4.2 A resolution | | Descriptor: | 1-methylpyrrolidine-2,5-dione, COPPER (II) ION, Plastocyanin | | Authors: | Cattani, G, Vogeley, L, Crowley, P.B. | | Deposit date: | 2014-08-01 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of a PEGylated protein reveals a highly porous double-helical assembly.
NAT.CHEM., 7, 2015
|
|
5CGB
 
 | | Crystal structure of FimH in complex with heptyl alpha-D-septanoside | | Descriptor: | (6R)-1,6-anhydro-2-O-heptyl-6-(hydroxymethyl)-D-galactitol, Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Preston, R.P, Zihlmann, P, Fiege, B, Sager, C.P, Vannam, R, Rabbani, S, Zalewski, A, Maier, T, Ernst, B, Peczuh, M. | | Deposit date: | 2015-07-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The price of flexibility - a case study on septanoses as pyranose mimetics.
Chem Sci, 9, 2018
|
|
4QVK
 
 | | Apo-crystal structure of Podospora anserina methyltransferase PaMTH1 | | Descriptor: | 1,2-ETHANEDIOL, PaMTH1 Methyltransferase | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
4QXT
 
 | | Crystal Structure of anti-MSP2 Fv fragment (mAb6D8)in complex with FC27-MSP2 14-30 | | Descriptor: | Fv fragment(mAb6D8) heavy chain, Fv fragment(mAb6D8) light chain, Merozoite surface antigen 2 | | Authors: | Morales, R.A.V, MacRaild, C.A, Seow, J, Bankala, K, Drinkwater, N, McGowan, S, Rouet, R, Christ, D, Anders, R.F, Norton, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for epitope masking and strain specificity of a conserved epitope in an intrinsically disordered malaria vaccine candidate.
Sci Rep, 5, 2015
|
|
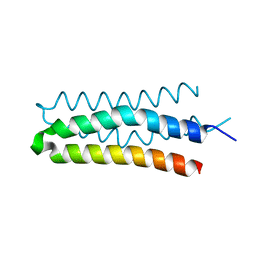
4R4L
 
 | | Crystal structure of wt cGMP dependent protein kinase I alpha (PKGI alpha) leucine zipper | | Descriptor: | HEXANE-1,6-DIOL, SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Reger, A.S, Guo, E, Yang, M.P, Qin, L, Kim, C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structures of cGMP-Dependent Protein Kinase (PKG) I alpha Leucine Zippers Reveal an Interchain Disulfide Bond Important for Dimer Stability.
Biochemistry, 54, 2015
|
|
5CP9
 
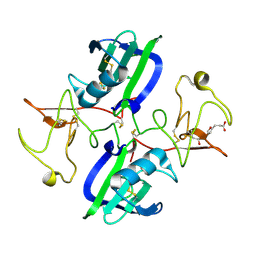 | | The structure of the NK1 fragment of HGF/SF complexed with MB605 | | Descriptor: | 1,2-ETHANEDIOL, 3-(furan-2-yl)propanoic acid, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
4RAV
 
 | | Crystal structure of scFvC4 in complex with the first 17 AA of huntingtin | | Descriptor: | Huntingtin, SULFATE ION, Single-chain Fv, ... | | Authors: | De Genst, E, Chirgadze, D.Y, Dobson, C.M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a single-chain fv bound to the 17 N-terminal residues of huntingtin provides insights into pathogenic amyloid formation and suppression.
J.Mol.Biol., 427, 2015
|
|
4QQB
 
 | | Structural basis for the assembly of the SXL-UNR translation regulatory complex | | Descriptor: | Protein sex-lethal, Upstream of N-ras, isoform A, ... | | Authors: | Hennig, J, Popowicz, G.M, Sattler, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the assembly of the Sxl-Unr translation regulatory complex.
Nature, 515, 2014
|
|
4TQG
 
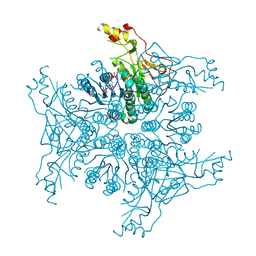 | | Crystal structure of Megavirus UDP-GlcNAc 4,6-dehydratase, 5-epimerase Mg534 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative dTDP-d-glucose 4 6-dehydratase | | Authors: | Jeudy, S, Piacente, F, De Castro, C, Molinaro, A, Salis, A, Damonte, G, Bernardi, C, Tonetti, M, Claverie, J.M, Abergel, C. | | Deposit date: | 2014-06-11 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Giant Virus Megavirus chilensis Encodes the Biosynthetic Pathway for Uncommon Acetamido Sugars.
J.Biol.Chem., 289, 2014
|
|
3RNM
 
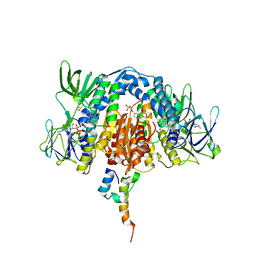 | | The crystal structure of the subunit binding of human dihydrolipoamide transacylase (E2b) bound to human dihydrolipoamide dehydrogenase (E3) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-MERCAPTOETHANOL, Dihydrolipoyl dehydrogenase, ... | | Authors: | Brautigam, C.A, Wynn, R.M, Chuang, J.C, Young, B.B, Chuang, D.T. | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Basis for Weak Interactions between Dihydrolipoamide Dehydrogenase and Subunit-binding Domain of the Branched-chain {alpha}-Ketoacid Dehydrogenase Complex.
J.Biol.Chem., 286, 2011
|
|
5E6C
 
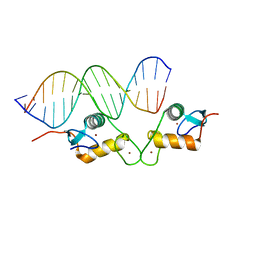 | | Glucocorticoid receptor DNA binding domain - CCL2 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
4RMW
 
 | |
5DZ5
 
 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P41212 space group | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
5E69
 
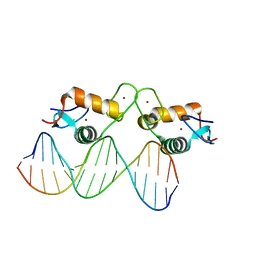 | | Glucocorticoid receptor DNA binding domain - IL8 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*CP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E6D
 
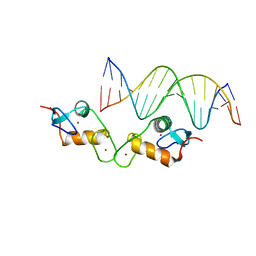 | | Glucocorticoid receptor DNA binding domain - ICAM1 NF-kB response element complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*GP*GP*AP*GP*C)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
4RL2
 
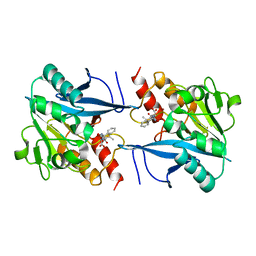 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (1R)-2-({(R)-carboxy[(2R,5S)-4-carboxy-5-methyl-5,6-dihydro-2H-1,3-thiazin-2-yl]methyl}amino)-2-oxo-1-phenylethanaminium, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4RM5
 
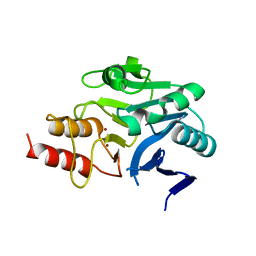 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
5DUN
 
 | | The crystal structure of OMe substituted twister ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (54-MER) | | Authors: | Ren, A, Patel, D.J, Micura, R, Rajashankar, K.R. | | Deposit date: | 2015-09-19 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
